SVD vs PCA vs 1bitMC
EigenDecomposition
For any real symmetric square d × d d \times d d×d matrix A , we can find its eigenvalues λ 1 ≥ λ 2 ≥ . . . ≥ λ d \lambda_1 \ge \lambda_2 \ge ... \ge \lambda_d λ1≥λ2≥...≥λd and corresponding orthonormal eigenvectors, such that:
A x 1 = λ 1 x 1 Ax_1 = \lambda_1x_1 Ax1=λ1x1
A
x
2
=
λ
1
x
2
Ax_2 = \lambda_1x_2
Ax2=λ1x2
…
A
x
d
=
λ
1
x
d
Ax_d = \lambda_1x_d
Axd=λ1xd
- Suppose there are only m different non-zero eigenvalues, so we have
A x 1 = λ 1 x 1 Ax_1 = \lambda_1x_1 Ax1=λ1x1
A
x
2
=
λ
1
x
2
Ax_2 = \lambda_1x_2
Ax2=λ1x2
…
A
x
r
=
λ
1
x
r
Ax_r = \lambda_1x_r
Axr=λ1xr
which can be written into a compact form such as ( U T = U − 1 U^T = U^{-1} UT=U−1 and ( U U T = I UU^T=I UUT=I):
A = U Σ U T (1) A=U\Sigma U^T \tag{1} A=UΣUT(1)
Σ
=
U
A
U
T
(2)
\Sigma=UAU^T \tag{2}
Σ=UAUT(2)
Then we use above matrix
A
A
A to tranform a vector x, then
A x = U Σ U T x = [ x 1 x 2 ⋯ x r ] [ λ 1 ⋱ λ r ] [ x 1 T x 2 T ⋯ x r T ] x \begin{aligned} Ax & =U\Sigma U^Tx \\ & = \left[ \begin{matrix} x_1 & x_2 \cdots & x_r \end{matrix} \right] \left[ \begin{matrix} \lambda_1 & & \\ & \ddots & \\ & & \lambda_r \end{matrix} \right] \left[ \begin{matrix} x_1^T \\ x_2^T\\ \cdots\\ x_r^T \end{matrix} \right] x \\ \end{aligned} Ax=UΣUTx=[x1x2⋯xr]⎣⎡λ1⋱λr⎦⎤⎣⎢⎢⎡x1Tx2T⋯xrT⎦⎥⎥⎤x
Start the calculation from right to left:
- Re-Orient: Apply U T U^T UT on x <==> dot proct of x with each rows of U T U^T UT. x will be projected on each orthonormal basis (new coordinate) which are the rows of the U T U^T UT. The geometric meaning of U T x U^Tx UTx is equivalent to rotate x based on the new cordinate (orthonormal basis) in U T U^T UT .
- Σ U T x \Sigma U^T x ΣUTx is to use Σ \Sigma Σ to extend or shrink the rotated x x x along with the new coordinate. (If there is one λ \lambda λ=0, then that dimension will be removed. )
- Re-Orient(to original):Same meaning with
U
T
U^T
UT.
U
U
U will rotate back x to original coordinate.

PCA
Main idea: Find a coordinate transformation matrix P applied on data suc that
(一种直观的看法是:希望投影后的投影值尽可能分散。数据越分散,可分性就越强,可分性越强,概率分布保存的就越完整)
-
The variability of the transformed data can be explained as much as possible along the new coordinates (降维后的信息损失尽可能小,尽可能保留原始样本的概率分布)
-
For each new coordinate, they should be orthoganal to each other to avoid the redundancy. (降维后的基之间是完全正交的)
-
So the final objective function is:
arg min P ∈ R m , d , U ∈ R d , m ∑ i = 1 n ∣ ∣ x i − U P x i ∣ ∣ 2 2 (*) \argmin_{P \in R^{m,d}, U\in R^{d,m}} \sum^{n}_{i=1} ||x_i - UPx_i||^2_2 \tag{*} P∈Rm,d,U∈Rd,margmini=1∑n∣∣xi−UPxi∣∣22(*)
, where d is the original dimension of data and m is the new dimension of the transoformed data (r=rank(A) <=m ), x 1 , . . . , x n ∈ R d x_1,..., x_n \in R^d x1,...,xn∈Rd , P ∈ R d , m P\in R^{d, m} P∈Rd,m is the transformation of new coordinate, U ∈ R d , m U \in R^{d,m} U∈Rd,m (in the end U = P T U = P^T U=PT)is the transoformation of original coordinate. -
In order to figure out above problems, the essential idea is to maxmize the variances of transfomred data point projected on new coordinates, and at the same time we need to minimize the covariance between two different coordinates(orthogonal<==>covarince is 0). Therefore, equivalently, we can diagolize the symmetric covariance matrix of the transformed data.
- Example (Let d =2).
Put data into matrix X:
X = [ x 1 , x 2 , . . . , x n ] = [ a 1 a 2 . . . a n b 1 b 2 . . . b n ] ∈ R d = 2 , n X=\left[ \begin{matrix} x_1, x_2, ...,x_n \end{matrix} \right] =\left[ \begin{matrix} a_1 & a_2 & ... &a_n\\ b_1 & b_2 & ... &b_n \end{matrix} \right] \in R^{d=2, n} X=[x1,x2,...,xn]=[a1b1a2b2......anbn]∈Rd=2,n
Suppose the data points has been centralized among each features. Therefore, the covariance matrix is the following:
A = 1 n X X T = [ 1 n ∑ i = 1 n a i 2 1 n ∑ i = 1 n a i b i 1 n ∑ i = 1 n a i b i 1 n ∑ i = 1 n b i 2 ] A=\frac{1}{n}XX^T= \left[ \begin{matrix} \frac{1}{n}\sum_{i=1}^{n} a^2_i & \frac{1}{n}\sum_{i=1}^{n} a_ib_i\\ \frac{1}{n}\sum_{i=1}^{n} a_ib_i\ & \frac{1}{n}\sum_{i=1}^{n} b^2_i \end{matrix} \right] A=n1XXT=[n1∑i=1nai2n1∑i=1naibi n1∑i=1naibin1∑i=1nbi2]
Transformed data by matrix P is Y = P X Y = PX Y=PX, where P ∈ R m , d = 2 P\in R^{m,d=2} P∈Rm,d=2. Then the transofrmed covariance matrix is:
1 n Y Y T = 1 n ( P X ) ( P X ) T = 1 n P X X T P T = P ( 1 n X X T ) P T = P A P T \begin{aligned} \frac{1}{n}YY^T & = \frac{1}{n}(PX)(PX)^T \\ & = \frac{1}{n}PXX^TP^T \\ & = P(\frac{1}{n}XX^T)P^T \\ & = PAP^T \\ \end{aligned} n1YYT=n1(PX)(PX)T=n1PXXTPT=P(n1XXT)PT=PAPT
Once the minimization (*) is done ,
1
n
Y
Y
T
\frac{1}{n}YY^T
n1YYT will look like
Σ
\Sigma
Σ. Underline technique used to achieve the goal is the formula (2) , so that
P
A
P
T
=
Σ
=
[
λ
1
⋱
λ
m
]
\begin{aligned} PAP^T & = \Sigma \\ & = \left[ \begin{matrix} \lambda_1 & & \\ & \ddots & \\ & & \lambda_m \end{matrix} \right] \end{aligned}
PAPT=Σ=⎣⎡λ1⋱λm⎦⎤
equivalently, A = P T Σ P A=P^T \Sigma P A=PTΣP
where λ 1 = σ 1 2 ≥ λ 2 = σ 2 2 ≥ . . . ≥ λ m = σ m 2 \lambda_1 =\sigma_1^2 \ge \lambda_2=\sigma_2^2 \ge ... \ge \lambda_m=\sigma_m^2 λ1=σ12≥λ2=σ22≥...≥λm=σm2, their corresponding eigenvectors construct the transformation matrix P = [ u 1 T u 2 T . . . u m T ] ∈ R m , d = 2 P=\left[ \begin{matrix} u_1^T\\ u_2^T\\ ...\\ u_m^T \end{matrix} \right] \in R^{m, d=2} P=⎣⎢⎢⎡u1Tu2T...umT⎦⎥⎥⎤∈Rm,d=2, depends on your problems, we can select different r<=m rows in P.

SVD
A
A
A will not be a symmetric matrix but with
m
×
n
m \times n
m×n dimension.
注意(SVD): Original Feature is n (the number of colmns), New Feature is m
注意(PCA above) : Original Feature is m (the number of rows)
Formula of Singular Value Decomposition
A
m
,
n
=
U
Σ
V
T
A_{m,n} = U \Sigma V^T
Am,n=UΣVT
The main word of SVD is trying to solve the following two tasks:
- First, find a set of orthonormal bais in n dimensional space
- Second, once performed on above space, the resulting basis in m dimensional space are still orthonormal.
Suppose we have already got the set of orthonormal bais in n dimensional space V n , n = [ v 1 , v 2 , . . . , v n ] V_{n,n} = \left[ v_1, v_2, ..., v_n \right] Vn,n=[v1,v2,...,vn] (note: some vs can be 0 vectors), where v i ⊥ v j v_i \perp v_j vi⊥vj
Project A on n dimensional bais, we have:
[
A
v
1
,
A
v
2
,
.
.
.
,
A
v
n
]
\left[ Av_1, Av_2, ..., Av_n \right]
[Av1,Av2,...,Avn]
-
To make the projected basis be orthogonal as well, we need let the following happen:
A v i ⋅ A v j = ( A v i ) T A v j = v i T A T A v j = 0 \begin{aligned} Av_i \cdot Av_j & = (Av_i)^T Av_j \\ & = v_i^T A^T A v_j \\ & = 0 \end{aligned} Avi⋅Avj=(Avi)TAvj=viTATAvj=0
Therefore, if v i s v_i s vis are eigen vectors of A T A A^TA ATA, then the resulting projected basis are also orthogonal because:
A v i ⋅ A v j = ( A v i ) T A v j = v i T ( A T A v j ) = v i T λ j v j = 0 \begin{aligned} Av_i \cdot Av_j & = (Av_i)^T Av_j \\ & = v_i^T (A^T A v_j )\\ & = v_i^T\lambda_j v_j \\ & = 0 \end{aligned} Avi⋅Avj=(Avi)TAvj=viT(ATAvj)=viTλjvj=0 -
To scale the projected orthogonal basis into unit length.
We have
u i = A v i ∣ A v i ∣ = A v i ∣ A v i ∣ 2 = A v i ( A v i ) T A v i ) = A v i v i T ( A T A v i ) = A v i v i T λ i v i ( n o t e : v i T v i = 1 ) = A v i λ i \begin{aligned} u_i & = \frac{ Av_i}{|Av_i|} \\ & = \frac{Av_i}{\sqrt{|Av_i|^2}} \\ & = \frac{Av_i}{\sqrt{(Av_i)^TAv_i)}} \\ & = \frac{Av_i}{\sqrt{v_i^T(A^TAv_i)}} \\ & = \frac{Av_i}{\sqrt{v_i^T\lambda_iv_i}} (note: v_i^Tv_i=1)\\ & = \frac{ Av_i}{\sqrt{\lambda_i}} \\ \end{aligned} ui=∣Avi∣Avi=∣Avi∣2Avi=(Avi)TAvi)Avi=viT(ATAvi)Avi=viTλiviAvi(note:viTvi=1)=λiAvi
u i λ i = u i σ i = A v i , u_i \sqrt{\lambda_i}= u_i \sigma_i = Av_i, uiλi=uiσi=Avi,, where σ i = λ i \sigma_i = \sqrt{\lambda_i} σi=λi is called singular value, 0 ≤ i ≤ r 0 \le i \le r 0≤i≤r, r=rank(A).
- In the end, we expand
[
u
1
,
u
2
,
.
.
.
,
u
r
]
\left[ u_1, u_2, ...,u_r\right]
[u1,u2,...,ur] to
[
u
1
,
u
2
,
.
.
.
,
u
r
∣
u
r
+
1
,
.
.
.
,
u
m
]
\left[ u_1, u_2, ...,u_r | u_{r+1}, ..., u_m\right]
[u1,u2,...,ur∣ur+1,...,um] and select
[
v
r
+
1
,
v
r
+
1
,
.
.
.
,
v
m
]
\left[ v_{r+1}, v_{r+1}, ...,v_{m}\right]
[vr+1,vr+1,...,vm] from the nullspace of A where
A
v
i
=
0
Av_i=0
Avi=0, i>r, and
σ
i
=
0
\sigma_i=0
σi=0
下图中k就是r

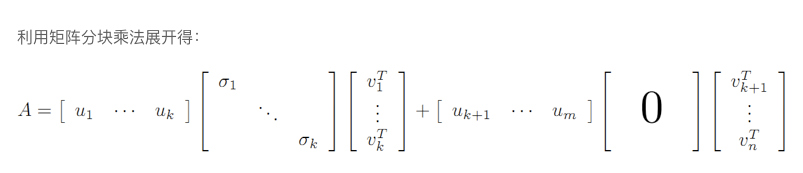


A = X Y A=XY A=XY
Take-away message
- The PCA just compute Left or Right Singular Matrix (depends how you define covariance matrix) of SVD.
- It’s unnecessary to construct a covariance matrix to start with SVD to decompose a matrix. In general, can use SGD.
Reference:
Ref1. (综合1+2)
Ref2. (1)
Ref3. (2)
























 2341
2341











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








