code:GitHub - VirtualFlow/VFVS: VirtualFlow for Virtual Screening
说明文档:GitBook
VirtualFlow平台旨在利用超级计算能力并行筛选潜在的有机化合物结构,以寻找有希望的新药物分子。
一种新药从开发到获得批准平均成本为20亿-30亿美元,至少耗时10年。这句话,药物研发领域的人大概都听累了。为什么这么难?
- 1. 湿实验昂贵而费时;
- 2. 初始化合物命中率低;
- 3. 临床前阶段的高损耗率。
今年3月,哈佛大学医学院(HMS)的研究人员在《Nature》杂志发表了论文《An open-source drug discovery platform enables ultra-large virtual screens》,描述了一个叫做VirtualFlow的开源药物发现平台,能通过云端整合海量的CPU对超大规模化合物库进行基于结构的虚拟筛选,提高药物发现效率。
论文作者Christoph Gorgulla称,在一个CPU上筛选10亿种化合物,每个配体的平均对接时间为15秒,全部筛完大概需要475年,而HMS利用VirtualFlow的平台,调用160000个CPU对接10亿个分子仅耗时约15小时,10000个CPU则需要两周。
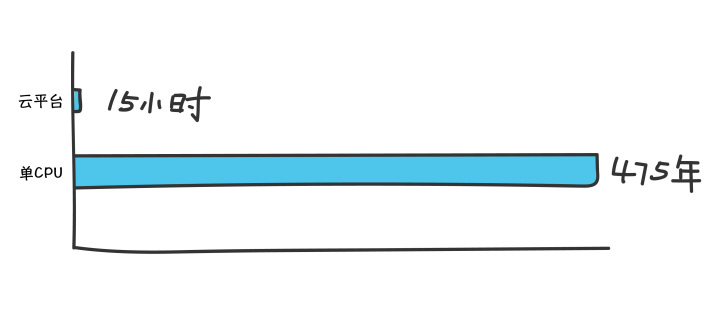
【听起来非常吸引人,抱着给某CRO公司虚拟筛选的7.8亿个分子,我们心里有点高兴,当时,我们调用了云上几万个core来筛选,计算时长也仅花费了3-13个小时(每个Core上所需时间不一样)】
限于算力,或者高效灵活地调用大规模计算集群的能力,当前的虚拟筛选通常仅采样百万到千万个分子,而事实上目前可用于药物发现的有机分子已经超过10的60次方。
注:湿实验室(Wet Lab)主要靠的是做实验,干实验室(Dry Lab)主要是计算机模拟和计算。
HMS的论文主要论证了两点:
1、虚拟筛选的规模越大,筛选的化合物越多,真阳性率越高;
2、线性扩展能力+云平台=无限可能。
超大规模筛选可提高真阳性率
论文推导了真阳性率与所筛选化合物数量的的函数关系的概率模型,证明:化合物的最高打分随着规模增加而提高。
作者分别从10万、100万、1000万、1亿、10亿个化合物中进行了5次筛选,挑选了得分最高的前50个化合物进行对比,从图中可以很清楚地看到筛选的规模越大,得分越高(位置越靠上)。

虚拟筛选规模可以通过两种不同的方式提高初始命中的质量:
1. 通过识别具有更紧密结合亲和力的化合物,从而降低剂量,减少脱靶效应;
2. 通过发现具有更好的药代动力学和/或更少固有细胞毒性的化合物。
为了验证大规模筛选的准确性,研究人员选择了肿瘤研究领域热门的KEAP1蛋白作为虚拟筛选靶点,对含有13亿配体的数据库进行了虚拟筛选。通过两个阶段的筛选,HMS选出了约1万个打分优秀的分子。随后,研究人员从成药性、配体效率、化学多样性以及获取难度等方面在这约1万个候选分子中挑选了590个苗头化合物进行活性验证,最终给出了两个活性达到毫微摩尔级的代表性化合物iKEAP 1和iKEAP 2的多种实验结果,验证了VirtualFlow在对接10亿以上分子量时的高效性。
线性扩展+云平台=无限可能
可线性扩展:处理器数量增加一倍,筛选能力也会增加一倍。
为了论证这一点,HMS在本地和云端均进行了测试:
本地计算集群LC1由18,000个CPU(分别为Intel Xeon和AMD Opteron处理器的不同型号)异构组成;本地集群LC2上则有30,000个英特尔Xeon8268处理器。
云端则选择了GCP和AWS,最多调用了160,000万个CPU(作者并未阐述在云端使用的CPU型号)。实验表明VirtualFlow在多种情况下均体现了良好的线性可扩展性,具体可看下图(图中并未描述平台在AWS上的表现)。
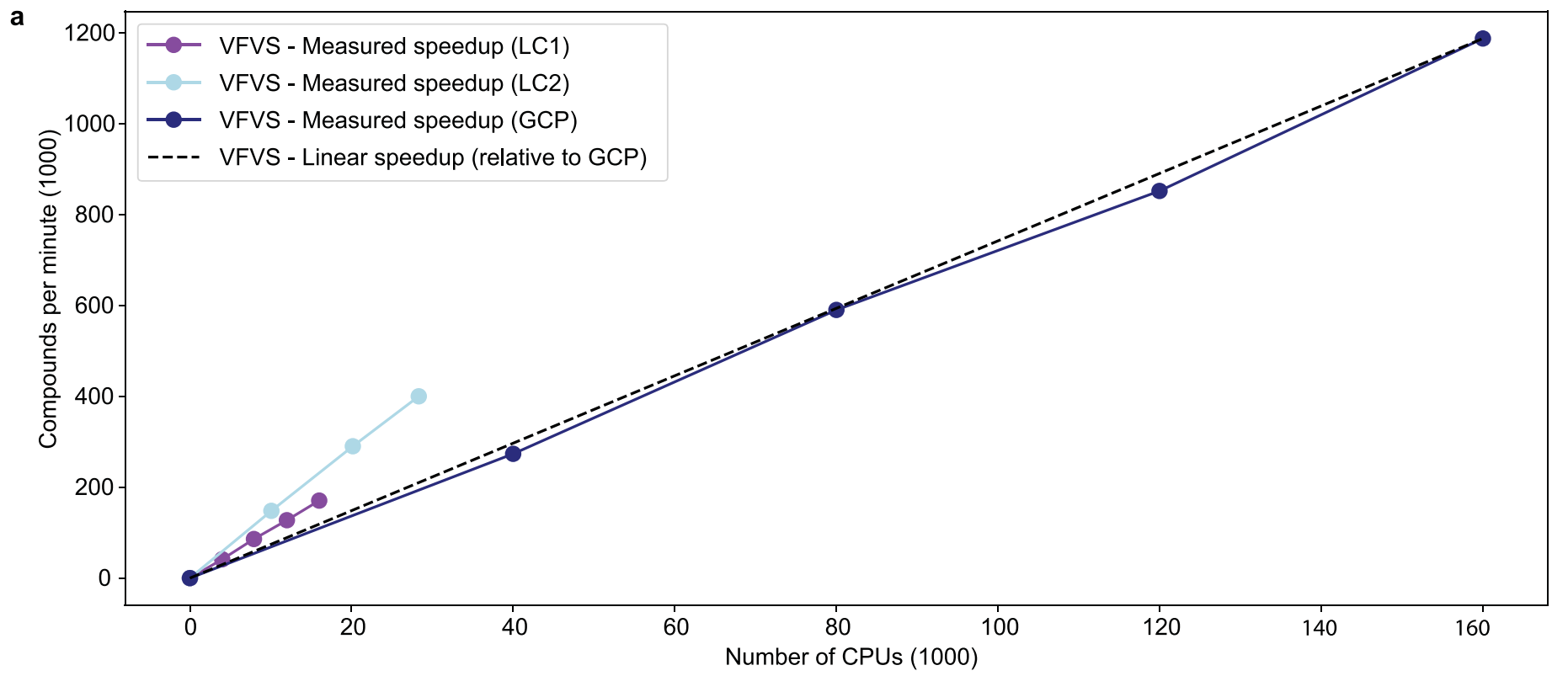
而这种近乎无限的线性扩展性意味着什么?
即便在今后的实际应用中并行数百万个内核,VirtualFlow的性能效率也不会受到其他因素的拖累。如果你拥有一个300核的计算机集群,你可以在六周内筛选1亿个化合物,而如果你有1,000核,那么两个星期内就可以完成筛选。
这个开源的VirtualFlow平台到底是个啥?
这个项目由哈佛大学医学院牵头,整体仍处于较新的阶段。VirtualFlow平台旨在利用超级计算能力并行筛选潜在的有机化合物结构,以寻找有希望的新药物分子。
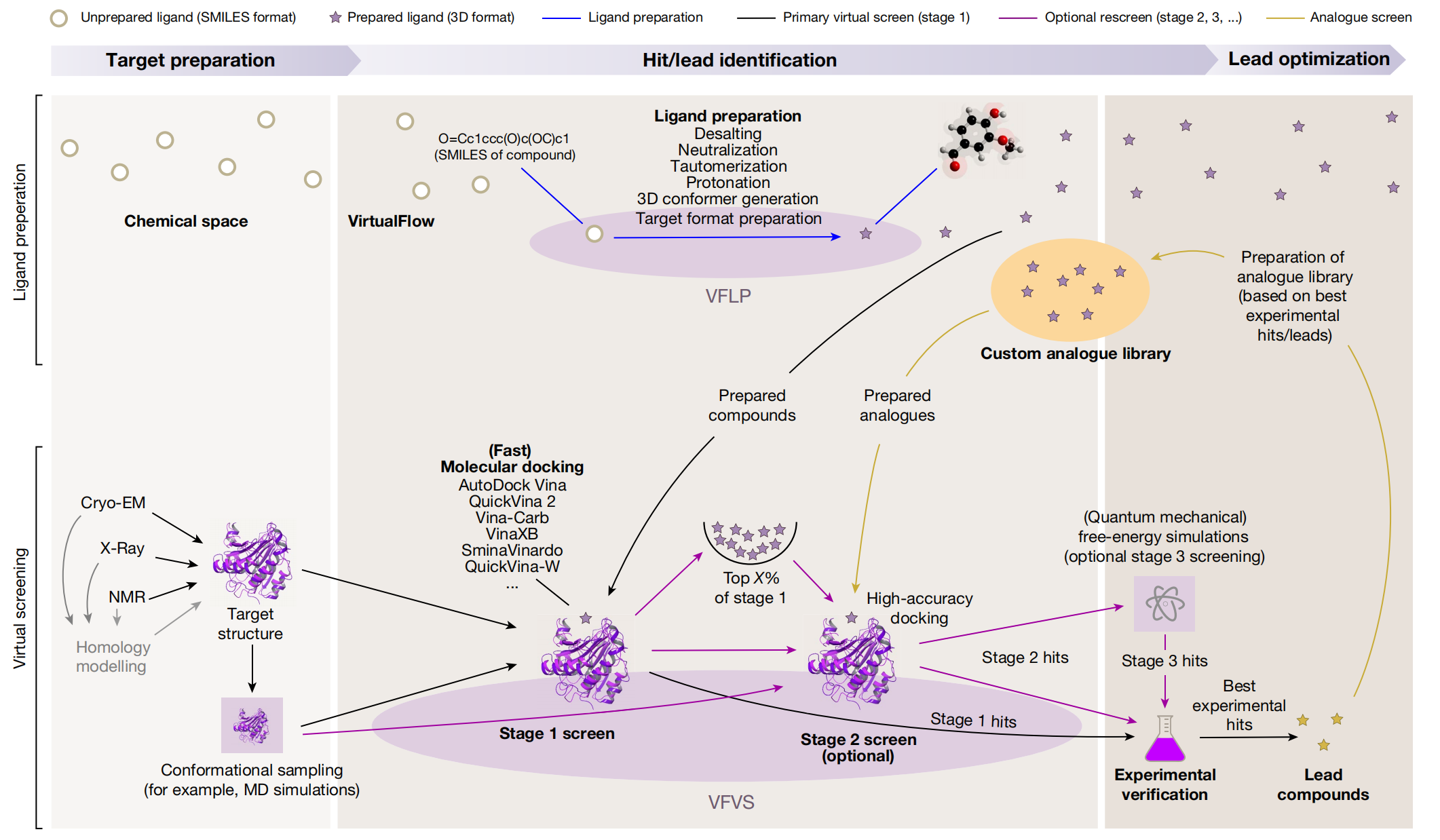
VirtualFlow平台主要分为VFLP(配体制备)和VFVS(虚拟筛选)两个模块,VFLP负责分析目标的化学空间构型(图中上半部分的蓝色箭头),再由VFVS根据事先预设好的靶点经过一次或多次虚拟筛选之后,最终获得先导化合物。
目前已知的平台特性包括:
1. 用Bash编写,完全开源、免费;
2. 目前支持的应用包括:AutoDock Vina、QuickVina 2、Smina、AutoDockFR、QuickVina-W、VinaXB和Vina-Carb;
3. 暂时不支持GPU;
4. 支持AWS、GCP、Azure在内的主流云计算平台。
15小时虚拟筛选10亿分子,《Nature》+HMS验证云端新药研发未来 - 速石科技 - 博客园
怎么运行起来?
1、首先需要安装slurm
https://medium.com/jacky-life/deploy-slurm-on-ubuntu-18-04-33abb6e902c4
----------------------------这里有一点一定要注意!!!!!!!!!!!!:------------------------
首先你先使用“sudo su”切换到root用户,再进行下面的操作,否则总会出很多的bug!!!
![]()
就是说下面的任务都是在root用户下进行的
----------------------------!!!!!!!!!!!!:------------------------
vim /etc/slurm-llnl/slurm.conf
根据:slurmd -C 指令了解这个node 的硬体配置:
NodeName=application-101 CPUs=64 Boards=1 SocketsPerBoard=2 CoresPerSocket=16 ThreadsPerCore=2 RealMemory=128847
UpTime=629-00:05:13
然后修改下面的内容, 主要是:“节点名字【application-101】、CPU数目【64】、内存【128847】一定要填对”
NodeName=application-101 CPUs=64 RealMemory=128847
# slurm.conf file generated by configurator easy.html.
# Put this file on all nodes of your cluster.
# See the slurm.conf man page for more information.
#
ControlMachine=application-101
#ControlAddr=
#
#MailProg=/bin/mail
MpiDefault=none
#MpiParams=ports=#-#
ProctrackType=proctrack/pgid
ReturnToService=1
SlurmctldPidFile=/var/run/slurm-llnl/slurmctld.pid
#SlurmctldPort=6817
SlurmdPidFile=/var/run/slurm-llnl/slurmd.pid
#SlurmdPort=6818
SlurmdSpoolDir=/var/spool/slurmd
SlurmUser=root
#SlurmdUser=root
StateSaveLocation=/var/spool/slurm-llnl
SwitchType=switch/none
TaskPlugin=task/none
#
#
# TIMERS
#KillWait=30
#MinJobAge=300
SlurmctldTimeout=3600
SlurmdTimeout=300
BatchStartTimeout=3600
PropagateResourceLimits=NONE
#
#
# SCHEDULING
FastSchedule=1
SchedulerType=sched/backfill
SelectType=select/linear
#SelectTypeParameters=
#
#
# LOGGING AND ACCOUNTING
#AccountingStorageType=accounting_storage/none
ClusterName=cluster
#JobAcctGatherFrequency=30
#JobAcctGatherType=jobacct_gather/none
#SlurmctldDebug=3
#SlurmctldLogFile=
#SlurmdDebug=3
#SlurmdLogFile=
#
# Acct
AccountingStorageEnforce=1
AccountingStorageLoc=/opt/slurm/acct
AccountingStorageType=accounting_storage/filetxt
JobCompLoc=/opt/slurm/jobcomp
JobCompType=jobcomp/filetxt
JobAcctGatherFrequency=30
JobAcctGatherType=jobacct_gather/linux
#
# COMPUTE NODES
NodeName=application-101 CPUs=64 RealMemory=128847 State=UNKNOWN
PartitionName=cpu101 Nodes=application-101 Default=YES MaxTime=INFINITE State=UP----------------------------!!!!!!!!!!!!:------------------------
注意:all.ctrl中的partition字段要和你的slume的partition一样才可以运行的起来:
通过sinfo查看当前的partition名称:

all.ctrl中的partition字段修改为此
partition=debug
----------------------------!!!!!!!!!!!!:------------------------
一些命令:【4.6】scancel取消运行或提交的作业 · Doc
首先是显示队列中所有的作业;
squeue
默认情况下squeue输出的内容如下,分别是作业号,分区,作业名,用户,作业状态,运行时间,节点数量,运行节点(如果还在排队则显示排队原因)
取消队列中已提交的作业;
介绍几个使用使用案例,分别是,取消指定作业、取消自己上机账号上所有作业、取消自己上机账号上所有状态为PENDING的作业,最后介绍scancel常见的参数。
取消指定作业;
# 取消作业ID为123的作业
scancel 123取消自己上机上号上所有作业;
# 注意whoami前后不是单引号
scancel -u `whoami`取消自己上机账号上所有状态为PENDING的作业;
scancel -t PENDING -u `whoami`
scancel常见参数;
--help # 显示scancel命令的使用帮助信息;
-A <account> # 取消指定账户的作业,如果没有指定job_id,将取消所有;
-n <job_name> # 取消指定作业名的作业;
-p <partition_name> # 取消指定分区的作业;
-q <qos> # 取消指定qos的作业;
-t <job_state_name> # 取消指定作态的作业,"PENDING", "RUNNING" 或 "SUSPENDED";
-u <user_name> # 取消指定用户下的作业;Slurm基础用法:
Slurm基础用法_muyuu的博客-CSDN博客_slurm srun
1.重启Slurm:
sudo systemctl restart slurmctld sudo systemctl restart slurmd.service
附:通过top命令查看当前CPU的占用

怎么运行该项目?
1、下载文件
上面的配置好以后,按照这个来就可以了:GitBook
The wget command can be used for downloading the file directly on the cluster:
wget https://virtual-flow.org/sites/virtual-flow.org/files/tutorials/VFVS_GK.tar
The tutorial files can now be extracted with the tar command:
tar -xvf VFVS_GK.tar
Going to the VirtualFlow Working Directory
At first, go to the folder to VFVS_GK/tools, since this is the working directory of VirtualFlow, where all commands are started:
cd VFVS_GK/tools
2、准备all.ctrl文件
Preparing the tools/templates/all.ctrl File
all.ctrl的配置:
*****************************************************************************************************************************************************************
*************************************************************** Job Resource Configuration ****************************************************************
*****************************************************************************************************************************************************************
job_letter=t
# One alphabetic character (i.e. a letter from a-z or A-Z)
# Should not be changed during runtime, and be the same for all joblines
# Required when running VF several times on the same cluster to distinguish the jobs in the batchsystem
# Settable via range control files: No
batchsystem=SLURM
# Possible values: SLURM, TOQRUE, PBS, LSF, SGE
# Settable via range control files: No
partition=cpu101
# Partitions are also called queues in some batchsystems
# Settable via range control files: Yes
timelimit=0-07:00:00
# Format for slurm: dd-hh:mm:ss
# Format for TORQUE and PBS: hh:mm:ss
# Format for SGE: hh:mm:ss
# Format for LSF: hh:mm
# For all batchsystems: always fill up with two digits per field (used be the job scripts)
# Settable via range control files: Yes
steps_per_job=1
# Not (yet) available for LSF and SGE (is always set to 1)
# Should not be changed during runtime, and be the same for all joblines
# Settable via range control files: Yes
cpus_per_step=64
# Sets the slurm cpus-per-task variable (task = step) in SLURM
# In LSF this corresponds to the number of slots per node
# Should not be changed during runtime, and be the same for all joblines
# Not yet available for SGE (always set to 1)
# Settable via range control files: Yes
queues_per_step=64
# Sets the number of queues/processes per step
# Should not be changed during runtime, and be the same for all joblines
# Not yet available for SGE (always set to 1)
# Settable via range control files: Yes
cpus_per_queue=1
# Should be equal or higher than <cpus-per-step/queues-per-step>
# Should not be changed during runtime, and be the same for all joblines
# Not yet available for SGE (always set to 1)
# Settable via range control files: Yes
*****************************************************************************************************************************************************************
********************************************************************* Workflow Options ********************************************************************
*****************************************************************************************************************************************************************
central_todo_list_splitting_size=1000
# When the folders are initially prepared the first time, the central todo list will be split into pieces of size <central_todo_list_splitting_size>. One task corresponds to one collection.
# Recommended value: < 100000, e.g. 10000
# Possible values: Positive integer
# The smaller the value, the faster the ligand collections can be distributed.
# For many types of clusters it is recommended if the total number of splitted todo lists stays below 10000.
# Settable via range control files: Yes
ligands_todo_per_queue=100
# Used as a limit of ligands for the to-do lists
# This value should be divisible by the next setting "ligands_todo_per_refilling_step"
# Settable via range control files: Yes
ligands_per_refilling_step=10
# The to-do files of the queues are filled with <ligands_per_refilling_step> ligands per refill step
# A number roughly equal to the average of number of ligands per collection is recommended
# Settable via range control files: Yes
collection_folder=../input-files/ligand-library
# Slash at the end is not required (optional)
# Relative pathname is required w.r.t. the folder tools/
# Settable via range control files: Yes
minimum_time_remaining=10
# In minutes
# A new job if the time left until the end of the walltime is smaller than the timelimit
# This is checked before each ligand is screened
# Thus the timelimit should be larger than the maximum time which is needed to process one ligand
# Settable via range control files: Yes
dispersion_time_min=3
# One positive integer, resembling the time in seconds
dispersion_time_max=10
# One positive integer, resembling the time in seconds
# The dispersion time is used when jobs try to access the central task list.
# Each job has to wait a random amount of time in the dispersion interval.
# The effect of this is that when two jobs arrive at the same time at the central task list, the random waiting time will disperse their access on the central task list in time
# Settable via range control files: Yes
verbosity_commands=standard
# Possible values: standard, debug
# This option mainly effects the screen output and the logfiles
# Settable via range control files: No
verbosity_logfiles=standard
# Possible values:
# * standard
# * debug : activates the set -x option. Increases size of log-files in average by nearly a factor of 10
# This option affects the preparation scripts for setting up the basic workflow files (before the workflow is running)
# Settable via range control files: Yes
store_queue_log_files=all_uncompressed
# Supported values (experimental)
# * all_uncompressed: requires most memory any and storage, but is recommending for test runs and debugging purposes
# * all_compressed: requires less memory and storage than uncompressed, but during the last part of the log files might get lost (in particular during crashes) due to the on-the-fly compression
# * only_error_uncompressed: only stderr is logged
# * only_error_compressed: only stderr is logged and compressed. The last part of the log might get lost (in particular during crashes) due to the on-the-fly compression.
# * std_compressed_error_uncompressed
# * all_compressed_error_uncompressed
# * none: reduces required memory and storage
# Settable via range control files: Yes
keep_ligand_summary_logs=true
# Summary log files which show for each ligand the success status of conversion and the conversion time.
# If the conversion failed, a reason is stated.
# If the transformation succeeded, the conversion programs which were used are stated.
# Possible values:
# * false
# * true
# Settable via range control files: Yes
error_sensitivity=normal
# Possible values: normal, high
# high sets the shell options "-uo pipefail". Not recommended for production runs, useful mainly for debugging. Pipefails often occur with tar combined with head/tail in pipes, which are not an actual problem.
# The u-option will always lead to a direct exit of the shell script when an unset variable is going to be used.
# Settable via range control files: Yes
error_response=fail
# Affects most errors, but not all (e.g. not the u-option of the shell)
# Possible values:
# * ignore : ignore error and continue
# * next_job : end this job and start new job
# * fail : exit workflow with failure (exit code 1)
# Settable via range control files: Yes
tempdir=/dev/shm
# The directory which is used for the temporary workflow files
# Should be a very fast filesystem not part of the distributed cluster filesystem, e.g. a local ramfs (as normally provided by ${VF_TMPDIR})
# The directory does only need to be available on the node on which the job step/queue is running
# In the tempdir, a subfolder named ${USER} will automatically be created
# Settable via range control files: Yes
*****************************************************************************************************************************************************************
***************************************************************** Virtual Screenign Options ***************************************************************
*****************************************************************************************************************************************************************
docking_scenario_names=qvina02_rigid_receptor1:smina_rigid_receptor1
# Values have to be separated by colons ":" and without spaces, e.g: docking_scenario_names=vina-rigid:smina-rigid
# Used for instance for the folder names in which the output files are stored
# Should not be changed during runtime, and be the same for all joblines
# Settable via range control files: Yes
docking_scenario_programs=qvina02:smina_rigid
# Possible values: qvina02, qvina_w, vina, smina_rigid, smina_flexible, adfr
# Values have to be separated by colons ":" and without spaces, e.g: docking_scenario_programs=vina:smina
# The same programs can be used multiple times
# Should not be changed during runtime, and be the same for all joblines
# Settable via range control files: Yes
docking_scenario_replicas=1:1
# Series of integers separated by colons ":"
# The number of values has to equal the number of docking programs specified in the variable "docking_programs"
# The values are in the same order as the docking programs specified in the variable "docking_scenario_programs
# e.g.: docking_scenario_replicas=1:1
# possible range: 1-99999 per field/docking program
# The docking scenario is comprised of all the docking types and their replicas
# Should not be changed during runtime, and be the same for all joblines
# Settable via range control files: Yes
docking_scenario_inputfolders=../input-files/qvina02_rigid_receptor1:../input-files/smina_rigid_receptor1
# Relative path wrt to the tools folders
# In each input folder must be the file config.txt which is used by the docking program to specify its options
# If other input files are required by the docking type, usually specified in the config.txt file, they have to be in the same folder
# Should not be changed during runtime, and be the same for all joblines
# Settable via range control files: Yes
*****************************************************************************************************************************************************************
******************************************************************* Terminating Variables *****************************************************************
*****************************************************************************************************************************************************************
stop_after_next_check_interval=false
# Determines whether the queue is stopped after the ligand batch currenty in progress. The size of the ligand batches is determined by the <ligand_check_interval> variable.
# Possible values:
# * false : The queue will continue to process ligands as long as there are ligands remaining for the queue
# * true : No new ligand will be started after the current ligand is completed
# Settable via range control files: Yes
ligand_check_interval=100
# Possible values
# * Positive integer
stop_after_collection=false
# Determines whether the queue is stopped after the current collection is completed
# Settable via range control files: Yes
# Possible values:
# * false : A new collection will be started if the current collection is completed and if there are collections remaining
# * true : No new collection will be started after the current collection is completed
# Settable via range control files: Yes
stop_after_job=false
# Determines whether the queue is stopped after the current job is completed
# Possible values:
# * false : A new job will be submitted if there are more ligands in the current collection or unprocessed collections remaining
# * true : No new job is submitted after the current job has ended
# Settable via range control files: Yes
其中最主要的是设置好这几项就可以了:
# 这个和你的slume的 “sinfo”命令中的PARTITION中的名字一样
partition=cpu101
# 每个作业 1 个节点(总共有多少个CPU就写多少)steps_per_job=1
# 每个节点使用多少个CPU核心线程数(如果是多线程的话),我这里是32核心64线程的机器cpus_per_step=64
# 和面的cpus_per_step的数目一致queues_per_step=64
# 一直为1cpus_per_queue=1
将 all.ctrl 文件放在/Virtual_Flow/VFVS_GK/tools/templates中,直接覆盖里面的 all.ctrl 就行
3、准备todo.all文件
配置文件信息都储存在all.crtl , todo.all中
todo.all中主要储存的是配体信息,按照下面的提示,等于cellections.txt中信息
如果你想要自己下载
a:下载配体文件库:
网站:https://virtual-flow.org/real-library
打开网站后,可以选择部分配体,也可以选择整个库下载,拖动图中紫色小球进行选择,本文选择的配体库如下图,点击Download,两个全部点击下载,本次选择wget

b:下载后的文件:
两个文件tranches.sh(第一个download),collections.txt(第二个download)

c:在终端下运行tranches,会下载一系列的文件
指令:sh tranches.sh 下载tranches中的EE文件,然后将该EE文件全部放入input-files的ligand-libary中

将collections.txt全部放入“VFVS_GK/tools/templates/”的todo.all文件中,也就是说将todo.all中的内容全部删除,然后将collections.txt中的内容全部放入
4、文件准备
./vf_prepare_folders.sh
5、运行
To start 12 jobs in parallel on a SLURM cluster, the following command can be used (within thetools directory):
./vf_start_jobline.sh 1 12 templates/template1.slurm.sh submit 1
由于这里我们只有一个CPU。所以选择命令为:
./vf_start_jobline.sh 1 1 templates/template1.slurm.sh submit 1
6、输出
To get preliminary virtual screening results for the docking scenario qvina02_rigid_receptor1, one can run the command (within the tools folder):
./vf_report.sh -c vs -d qvina02_rigid_receptor1 -n 10
The -n option here specifies that we would also like to have the top 10 compounds listed.
输出的结果在output-files文件夹中
附:如果你成功使用了所有核心,那么应该是CPU占用率接近百分之百,然后每个核心都占用为100(top 命令):

==============一路上太多的坑,如果对你有帮助,那这篇文章就实现了它的价值 !================
office 指导文件:https://docs.virtual-flow.org/tutorials/-LdE94b2AVfBFT72zK-v/vfvs-tutorial-1-bash/installation
其他指导文件:Nature | 手把手教你搭建大规模药物虚拟筛选平台
VirtualFlow教程 | Enamine数据库的更新及其虚拟筛选
python对csv文件中的数据进行筛选_Nature | 手把手教你搭建大规模药物虚拟筛选平台..._weixin_39588265的博客-CSDN博客
http://blog.molcalx.com.cn/2020/07/20/cloudam-virtualflow-htvs.html


























 2075
2075

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










