1.输入文件,gff3文件
2.脚本:windows版本
windows版本
import csv
def parse_gff3(input_file):
features = {}
with open(input_file, 'r', encoding='utf-8') as file: # 指定文件编码
reader = csv.reader(file, delimiter='\t')
for row in reader:
if row[0].startswith('#') or len(row) < 9:
continue
seqid, source, feature_type, start, end, score, strand, phase, attributes = row
attr_dict = {attr.split('=')[0]: attr.split('=')[1] for attr in attributes.split(';') if '=' in attr}
if feature_type in ['five_prime_UTR', 'CDS', 'three_prime_UTR']:
parent_id = attr_dict.get('Parent')
if parent_id in features:
if feature_type not in features[parent_id]:
features[parent_id][feature_type] = []
features[parent_id][feature_type].append({'start': int(start), 'end': int(end)})
else:
features[parent_id] = {feature_type: [{'start': int(start), 'end': int(end)}]}
return features
def write_new_gff3(features, output_file):
with open(output_file, 'w', encoding='utf-8', newline='') as file: # 避免 Windows 下的空行问题
file.write("ID\tType\tStart\tEnd\tLength\n")
for transcript_id, data in features.items():
position = 1
for feature_type in ['five_prime_UTR', 'CDS', 'three_prime_UTR']:
if feature_type in data:
combined_coords = sorted(data[feature_type], key=lambda x: x['start'])
total_length = sum(coord['end'] - coord['start'] + 1 for coord in combined_coords)
new_end = position + total_length - 1
file.write(f"{transcript_id}\t{feature_type}\t{position}\t{new_end}\t{total_length}\n")
position = new_end + 1
# Hardcoded file paths
input_file = 'E:/pythonworking/file/20240430.txt'
output_file = 'E:/pythonworking/file/20240430out.txt'
features = parse_gff3(input_file)
write_new_gff3(features, output_file)
linux版本:
import csv
import argparse
def parse_gff3(input_file):
""" Read a GFF3 file and parse 5'UTR, CDS, and 3'UTR features of transcripts """
features = {}
with open(input_file, 'r') as file:
reader = csv.reader(file, delimiter='\t')
for row in reader:
if row[0].startswith('#') or len(row) < 9:
continue
seqid, source, feature_type, start, end, score, strand, phase, attributes = row
attr_dict = {attr.split('=')[0]: attr.split('=')[1] for attr in attributes.split(';') if '=' in attr}
if feature_type in ['five_prime_UTR', 'CDS', 'three_prime_UTR']:
parent_id = attr_dict.get('Parent')
if parent_id in features:
if feature_type not in features[parent_id]:
features[parent_id][feature_type] = []
features[parent_id][feature_type].append({
'start': int(start),
'end': int(end)
})
else:
features[parent_id] = {feature_type: [{'start': int(start), 'end': int(end)}]}
return features
def write_new_gff3(features, output_file):
""" Write a new GFF3 file based on parsed features, including ID, Type, Start, End, and Length """
with open(output_file, 'w') as file:
file.write("ID\tType\tStart\tEnd\tLength\n")
for transcript_id, data in features.items():
position = 1
for feature_type in ['five_prime_UTR', 'CDS', 'three_prime_UTR']:
if feature_type in data:
coords_list = data[feature_type]
# Combine all coordinates and sort them
combined_coords = sorted(coords_list, key=lambda x: x['start'])
# Calculate the total length of combined segments
total_length = sum(coord['end'] - coord['start'] + 1 for coord in combined_coords)
new_end = position + total_length - 1
file.write(f"{transcript_id}\t{feature_type}\t{position}\t{new_end}\t{total_length}\n")
position = new_end + 1 # Update the position for the next feature
def main():
parser = argparse.ArgumentParser(description="Reformat GFF3 file to transcript-centric simplified format with additional length info")
parser.add_argument("-i", "--input", required=True, help="Input GFF3 file path")
parser.add_argument("-o", "--output", required=True, help="Output GFF3 file path")
args = parser.parse_args()
# Parse the GFF3 file
features = parse_gff3(args.input)
# Output the new format GFF3 file
write_new_gff3(features, args.output)
if __name__ == "__main__":
main()
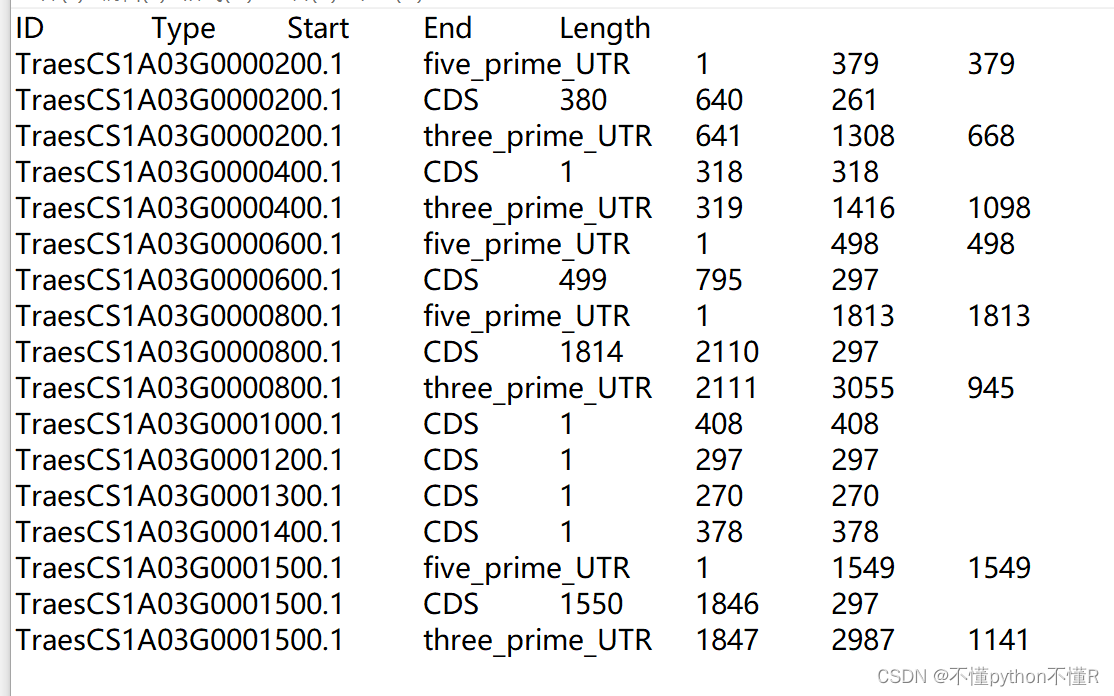
3.输出文件






















 776
776











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








