欢迎大家关注全网生信学习者系列:
- WX公zhong号:生信学习者
- Xiao hong书:生信学习者
- 知hu:生信学习者
- CDSN:生信学习者2
介绍
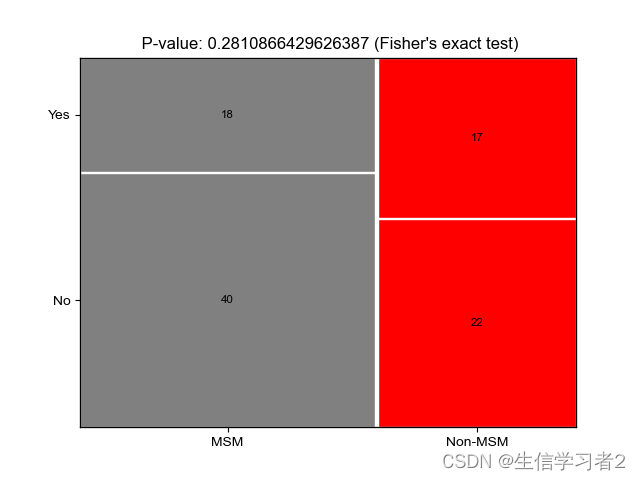
本教程是使用一个Python脚本来绘制马赛克图,用于可视化两个变量的频率分布。
数据
大家通过以下链接下载数据:
- 百度网盘链接:https://pan.baidu.com/s/1f1SyyvRfpNVO3sLYEblz1A
- 提取码: 请关注WX公zhong号_生信学习者_后台发送 复现msm 获取提取码
Python packages required
Drawing a mosaic plot using mosaic_plot.py
使用一个Python脚本mosaic_plot.py,以及一个包含MSM 和 Non-MSM个体相关的物种的表格,这些物种被识别为革兰氏阴性或非革兰氏阴性,在two_variable_mosaic.tsv: ./data/two_variable_mosaic.tsv中。
mosaic_plot.pycodes
#!/usr/bin/env python
"""
NAME: mosaic_plot.py
DESCRIPTION: mosaic_plot.py is a python script for visualizing proportions of data points along two variables.
"""
import pandas as pd
from scipy.stats import fisher_exact
import matplotlib.pyplot as plt
from statsmodels.graphics.mosaicplot import mosaic
import matplotlib
import sys
import argparse
import textwrap
def make_mosaic_plot(two_variable_file, facecolor_dict, output_fig, font_style = "sans-serif,Arial"):
font_family, font_type = font_style.split(",")
matplotlib.rcParams['font.family'] = font_family
matplotlib.rcParams['font.sans-serif'] = font_type
two_variable_df = pd.read_csv(two_variable_file, sep = "\t", index_col = False)
features, variable1, variable2 = two_variable_df.columns
cont_df = pd.crosstab(two_variable_df[variable1], two_variable_df[variable2])
res = fisher_exact(cont_df, alternative = "two-sided")
label_dict = {}
for idx in cont_df.index.to_list():
for col in cont_df.columns.to_list():
label_dict[(idx, col)] = cont_df.loc[idx, col]
labelizer = lambda k:label_dict[k]
variable2_0, variable2_1 = sorted(set(two_variable_df[variable2].to_list()))
props = {}
for variable in facecolor_dict:
props[(variable, variable2_0)] = {"facecolor": facecolor_dict[variable], "edgecolor": "white"}
props[(variable, variable2_1)] = {"facecolor": facecolor_dict[variable], "edgecolor": "white"}
mosaic(two_variable_df, [variable1, variable2], labelizer = labelizer, properties = props, title = " P-value: "+ str(res[1]) + " (Fisher's exact test)")
plt.savefig(output_fig)
if __name__ == "__main__":
def read_args(args):
# This function is to parse arguments
parser = argparse.ArgumentParser(formatter_class=argparse.RawDescriptionHelpFormatter,
description = textwrap.dedent('''\
This program is to draw a mosaic plot.
'''),
epilog = textwrap.dedent('''\
examples: mosaic_plot.py --input input_file.tsv --facecolor_map facecolor_mapfile.tsv --output mosaic_plot.png
'''))
parser.add_argument('--input',
nargs = '?',
help = 'Input a file containing two variable information regarding each individual subject.',
type = str,
default = None)
parser.add_argument('--facecolor_map',
nargs = '?',
help = 'Specify the the pathway to SCFA metabolisms database. default: /vol/projects/khuang/databases/SCFA/SCFA_pathways.tsv',
default = '/vol/projects/khuang/databases/SCFA/SCFA_pathways.tsv')
parser.add_argument('--font_style',
nargs = '?',
help = 'Specify the font style, font family and font type is delimited by a comma. default: [sans-serif,Arial]',
default = 'sans-serif,Arial')
parser.add_argument('--output',
nargs = '?',
help = 'Specify the output figure name.',
type = str,
default = None)
return vars(parser.parse_args())
pars = read_args(sys.argv)
facecolor_dict = {i.rstrip().split("\t")[0]: i.rstrip().split("\t")[1] for i in open(pars['facecolor_map']).readlines()}
make_mosaic_plot(pars["input"], facecolor_dict , pars["output"], font_style = pars["font_style"])
- Usage:
mosaic_plot.py [-h] [--input [INPUT]] [--facecolor_map [FACECOLOR_MAP]] [--font_style [FONT_STYLE]] [--output [OUTPUT]]
This program is to draw a mosaic plot.
optional arguments:
-h, --help show this help message and exit
--input [INPUT] Input a file containing two variable information regarding each individual subject.
--facecolor_map [FACECOLOR_MAP]
Specify the the pathway to SCFA metabolisms database. default: /vol/projects/khuang/databases/SCFA/SCFA_pathways.tsv
--font_style [FONT_STYLE]
Specify the font style, font family and font type is delimited by a comma. default: [sans-serif,Arial]
--output [OUTPUT] Specify the output figure name.
examples:
python mosaic_plot.py --input input_file.tsv --facecolor_map facecolor_mapfile.tsv --output mosaic_plot.png
示例命令:
python mosaic_plot.py \
--input two_variable_mosaic.tsv \
--facecolor_map facecolor_map.tsv \
--output mosaic_plot.png
Note
马赛克图的面颜色应该按照示例中的映射文件mapping file: ./data/facecolor_map.tsv来指定。























 157
157

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










