做过克隆构建的人都知道,有时候我们需要对测序结果进行分析,查看测序结果是否与我们想要构建的序列是否一致,即使是构建同一条序列的克隆,有时候根据实验实际情况,我们需要测序的单克隆不止两三个,有可能是五六个,也有可能是几十上百个,面对大量的测序结果,如果我们进行一一翻译比对,那么无疑花费我们大量时间,耗费不少精力,下面展示如何通过代码进行批量的处理并输出结果。
一、读取测序数据并进行相关设置
import os
path = "E:\gene_counts"
path1 = "E:\gene_counts"
head = "CCATGG" #输入起始翻译位置
tail = "TIEWA" #输入目的序列尾部氨基酸
klyk = "MVQKSLLFSLLASTALGALTKRYSFPLPESQGSETFSEPYEVAAGETFDGGMKTYGRGVECTGQVEGGEDDTVFIVQEGGTLKNAIIGTDQIEGVYCLGSCTIENVWWEKVCEDALSLKEGDGIYTISGGGAQGAEDKVIQHNTGGEVIIDGFEVYDFGKLYRSCGTCGDIQRKVSVSNVVAVSGSQLVGINENFGDTATIDSSVCATDVNDICATYNGTDGDEEPEEVSTGPSDYCIYTEPIAECA"
#输入想要比对的目的序列二、将合格测序数据整理打包
正向(5‘端到3‘端测序)测序数据处理:
for a in os.walk(path):
a = a[2]
f1 = open(path1 + "\测序结果生成.txt", "w")
for b in a:
if b[-8:] == "[T7].seq":
f = open(path + "/" + b, "r")
for c in f.readlines():
d = c
if head in d:
e = d.index(head)
m = d[e + 2:]
f1.write(">" + b[21:-10] + "\n" + m + "\n")
f.close()
f1.write(">" + "\n")
f1.close()
反向(3‘端到5‘端测序)测序数据处理:
import os
for a in os.walk(path):
a = a[2]
f1 = open(path1 + "/测序结果生成.txt", "w")
for b in a:
if b[-12:] == "[T7-TER].seq":
f = open(path + "/" + b, "r")
for c in f.readlines():
d = c
if head in d:
e = d.index(head)
m = d[0:e + 4]
f1.write(">" + b[21:-14] + "\n" + m + "\n")
f.close()
f1.write(">" + "\n")
f1.close()正向测序结果和反向测序数据拼接:
import os
for a in os.walk(path):
a = a[2]
zheng = {}
for b in a:
if b[-8:] == "[T7].seq":
f = open(path + "/" + b, "r")
for c in f.readlines():
d = c
if head in d:
e = d.index(head)
m = d[e + 2:]
zheng.update({">" + b[21:-10]:m})
f.close()
fan = {}
for b in a:
if b[-12:] == "[T7-TER].seq":
f = open(path + "/" + b, "r")
for c in f.readlines():
d = c
if head in d:
e = d.index(head)
m = d[0:e + 4]
mw = ""
for n in m:
if n == "A":
n = "T"
mw += n
elif n == "T":
n = "A"
mw += n
elif n == "G":
n = "C"
mw += n
elif n == "C":
n = "G"
mw += n
fan.update({">" + b[21:-14]:mw[::-1]})
f.close()
f1 = open(path1 + "/测序结果生成.txt", "w")
for key in zheng:
if key in fan:
kk = zheng[key][-200:-190]
if kk in fan[key]:
mm = fan[key][fan[key].index(kk):]
yy = zheng[key][0:-200]
nn = yy + mm
f1.write(key + "\n" + nn + "\n")
f1.write(">" + "\n")
f1.close()
三、将翻译的数据进行清洗打包
#在线网站进行一键翻译https://www.novopro.cn/tools/translate.html
f = open(path1 + "/测序结果生成.txt", "r")
b = ""
diction1 = []
diction2 = []
i = 0
for a in f.readlines():
i += 1
if i == 1:
name = a[5:-1]
else:
if a[0] != ">":
b += a.replace("\n", "")
if a[0] == ">":
if tail in b:
diction1.append((name,b))
diction2.append((name,b[0:len(ClyC) + 9]))
print((name,b[0:(len(ClyC) + 9)]))
b = ""
name = a[5:-1]
f.close()
四、寻找并输出突变位点(不突变输出None)
for x, y in diction2:
if tail in y:
if len(y[0:(y.index(tail) + len(tail))]) == len(ClyC):
print(y)
if y[0:(y.index(tail) + len(tail))] == ClyC:
print(x, "None")
else:
for b in range(len(ClyC)):
if ClyC[b] != y[b]:
print(x, ":", ClyC[b] + str(b + 1) + y[b])
elif len(y[0:(y.index(tail) + len(tail))]) > len(ClyC):
print(y + "\n" + x , ":插入突变")
elif len(y[0:(y.index(tail) + len(tail))]) < len(ClyC):
print(y + "\n" + x , ":缺失突变")
五、输出结果展示
MVQKSLLFSLLASTALGALTKRYSFPLPESQGSETFSEPYEVAAGETFDGGMKTYGRGVECTGQVEGGEDDTVFIVQEGGTLKNAIIGTDQIEGVYCLGSCTIENVWWEKVCEDALSLKEGDGIYTISGGGAQGAEDKVIQHNTGGEVIIDGFEVYDFGKLYRSCGTCGDIQRKVSVSNVVAVSGSQLVGINENFGDTATIDSSVCATDVNDICATYNGTDGDEEPEEVSTGPSDYCIYTEPIAECA
3-3 : P81K
3-3 : S170I
MVQKSLLFSLLASTALGALTKRYSFPLPESQGSETFSEPYEVAAGETFDGGMKTYGRGVECTGQVEGGEDDTVFIVQEGGTLKNAIIGTDQIEGVYCLGSCTIENVWWEKVCEDALSLKEGDGIYTISGGGAQGAEDKVIQHNTGGEVIIDGFEVYDFGKLYRSCGTCGDIQRKVSVSNVVAVSGSQLVGINENFGDTATIDSSVCATDVNDICATYNGTDGDEEPEEVSTGPSDYCIYTEPIAECA
7-3 : Y209K
MVQKSLLFSLLASTALGALTKRYSFPLPESQGSETFSEPYEVAAGETFDGGMKTYGRGVECTGQVEGGEDDTVFIVQEGGTLKNAIIGTDQIEGVYCLGSCTIENVWWEKVCEDALSLKEGDGIYTISGGGAQGAEDKVIQHNTGGEVIIDGFEVYDFGKLYRSCGTCGDIQRKVSVSNVVAVSGSQLVGINENFGDTATIDSSVCATDVNDICATYNGTDGDEEPEEVSTGPSDYCIYTEPIAECA
9-3 : W19C
9-3 : W177L
9-3 : T251K
MVQKSLLFSLLASTALGALTKRYSFPLPESQGSETFSEPYEVAAGETFDGGMKTYGRGVECTGQVEGGEDDTVFIVQEGGTLKNAIIGTDQIEGVYCLGSCTIENVWWEKVCEDALSLKEGDGIYTISGGGAQGAEDKVIQHNTGGEVIIDGFEVYDFGKLYRSCGTCGDIQRKVSVSNVVAVSGSQLVGINENFGDTATIDSSVCATDVNDICATYNGTDGDEEPEEVSTGPSDYCIYTEPIAECA
9-4 : None
MVQKSLLFSLLASTALGALTKRYSFPLPESQGSETFSEPYEVAAGETFDGGMKTYGRGVECTGQVEGGEDDTVFIVQEGGTLKNAIIGTDQIEGVYCLGSCTIENVWWEKVCEDALSLKEGDGIYTISGGGAQGAEDKVIQHNTGGEVIIDGFEVYDFGKLYRSCGTCGDIQRKVSVSNVVAVSGSQLVGINENFGDTATIDSSVCATDVNDICATYNGTDGDEEPEEVSTGPSDYCIYTEPIAECA
9-2 : 插入突变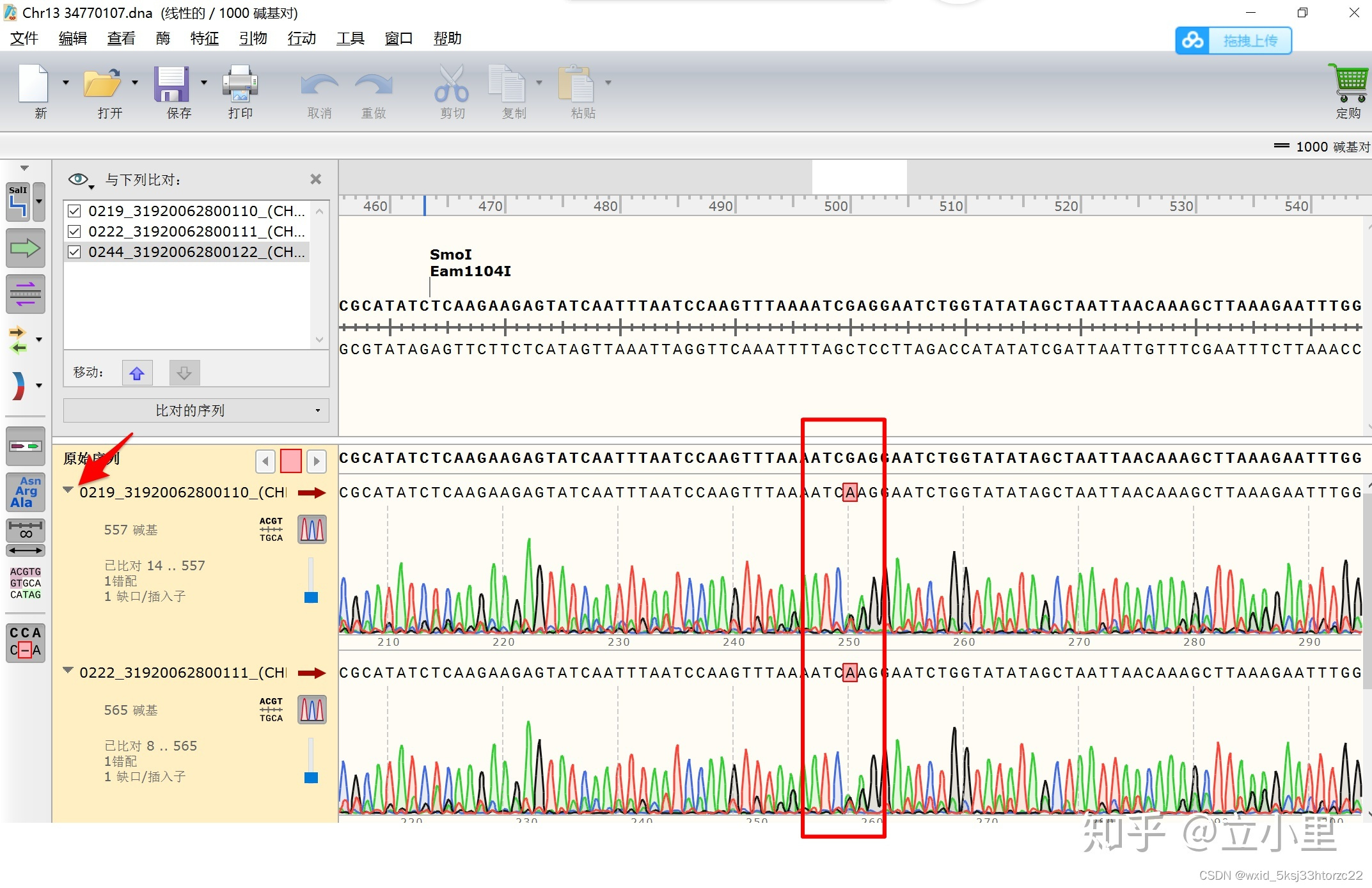
如何对构建基因克隆的测序结果批量进行序列比对并输出结果 (qq.com)
感兴趣点赞收藏或者转发,谢谢大家!







 文章介绍了一种通过Python代码自动化处理大量基因克隆测序数据的方法,包括读取正反向测序数据、拼接比对以及查找突变位点,从而节省手动比对的时间和精力。
文章介绍了一种通过Python代码自动化处理大量基因克隆测序数据的方法,包括读取正反向测序数据、拼接比对以及查找突变位点,从而节省手动比对的时间和精力。

















 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








