
“Sklearn源码机器学习笔记” 系列教程以Scikit-learn 0.19.x库为基础,系列笔记从Sklearn包源码入手进行理解算法,同时对于sklearn文档的部分demo进行深入的学习。
由于能力有限,不足支持请大家多多指正,大家有什么想法也非常欢迎留言评论!
关于我的更多学习笔记,欢迎您关注“武汉AI算法研习”公众号,Sklearn官网最新原版英文文档下载,公众号回复“sklearn”。
本文分三个部分“【高斯朴素贝叶斯】”、“【多项式朴素贝叶斯】”、“【伯努利朴素贝叶斯】”来进行展开,总共阅读时间大约10分钟。
关于朴素贝叶斯原理方法文章,可见公众号的文章“李航《统计学习方法》学习笔记之——第九章:EM算法及其推广(高斯混合模型)”
Sklearn下关于高斯混合模型位于mixture/gaussian_mixture.py文件,同目录下也存在mixture/bayesian_mixture.py文件,
#GuassianMixture构造函数方法:
class sklearn.mixture.GaussianMixture(n_components=1, covariance_type=’full’, tol=0.001, reg_covar=1e-06, max_iter=100, n_init=1, init_params=’kmeans’, weights_init=None, means_init=None, precisions_init=None, random_state=None, warm_start=False, verbose=0, verbose_interval=10)[source]# Sklearn关于高斯混合模型的使用demo
# fit a Gaussian Mixture Model with two components
clf = mixture.GaussianMixture(n_components=2, covariance_type='full')
clf.fit(X_train)在官方的Sklearn的官方文档中,关于高斯混合模型的Examples例子总共有6个,通过6个例子可以很好了解混合高斯模型的工程运用。
- 高斯分布的概率密度估计
- 高斯模型选择
- 高斯协方差
- 用 GaussianMixture 和 BayesianGaussianMixture 绘制置信椭圆体
- 选择经典高斯混合模型中分量的个数
- 浓度先验型变异贝叶斯高斯混合分析
【Density Estimation for a Gaussian mixture】
Plot the density estimation of a mixture of two Gaussians. Data is generated from two Gaussians with different centers and covariance matrices.
官网样例中训练样本数为600,样本特征维度为2,高斯混合模型的分模型数为2,样例代码分别生成两个以(20,20)和(0,0)为中心的高斯数据,通过600个训练样本调用高斯混合模型,进行训练得到高斯混合模型。图例中红色点表训练模型的600个样本数据,图中黄色点表绘制高斯分布等概率密度数据点,绘制登高线过程中contour()函数根据均匀分布的X和Y坐标,计算每个坐标点的概率密度值,最后经过一定的平滑和等高线间距生成等高线图,从图中可以知道等高线形状这个呈现出椭圆。
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import LogNorm
from sklearn import mixture
n_samples = 300
np.random.seed(0)
# 生成球形数据集,数据中心为(20,20)
# randn()返回300 * 2 的具有标准正态分布的数据,同时样本数据点增加20,以此(20, 20)为样本中心。
shifted_gaussian = np.random.randn(n_samples, 2) + np.array([20, 20])
# 生成以(0,0)为中心的高斯数据,
# dot()返回的是两个数组的点积
C = np.array([[0., -0.7], [3.5, .7]])
stretched_gaussian = np.dot(np.random.randn(n_samples, 2), C)
# np.vstack:按垂直方向(行顺序)堆叠数组构成一个新的数组
X_train = np.vstack([shifted_gaussian, stretched_gaussian])
# 训练高斯混合模型
clf = mixture.GaussianMixture(n_components=2, covariance_type='full')
clf.fit(X_train)
# 产生测试数据
x = np.linspace(-20., 30.) # linspace()指定的间隔内返回均匀间隔的数字。
y = np.linspace(-20., 40.)
X, Y = np.meshgrid(x, y) # np.meshgrid()—生成网格点坐标矩阵
XX = np.array([X.ravel(), Y.ravel()]).T #ravel():多维数组转换为一维数组,如果没有必要,不会产生源数据的副本
dd = clf.predict(XX)
Z = -clf.score_samples(XX)
Z = Z.reshape(X.shape)
CS = plt.contour(X, Y, Z, norm=LogNorm(vmin=1.0, vmax=1000.0),
levels=np.logspace(0, 3, 10))
CB = plt.colorbar(CS, shrink=0.8, extend='both')
plt.scatter(X_train[:, 0], X_train[:, 1], .8, edgecolors = [1, 0, 0] ) # scatter()画散点图
plt.scatter(XX[:, 0], XX[:, 1], .8) # scatter()画散点图
plt.title('Negative log-likelihood predicted by a GMM')
plt.axis('tight')
plt.show()

【Gaussian Mixture Model Ellipsoids】
Both models have access to five components with which to fit the data. Note that the Expectation Maximisation model will necessarily use all five components while the Variational Inference model will effectively only use as many as are needed for a good fit. Here we can see that the Expectation Maximisation model splits some components arbitrarily, because it is trying to fit too many components, while the Dirichlet Process model adapts it number of state automatically.
GaussianMixture用EM算法拟合数据,调用函数中k等于几就拟合出几个分量,人为控制拟合几个分高斯模型。BayesianGaussianMixture 用了 Variational Inference model(变分推断模型)可以自适应最佳的簇数,即便制定了components=5,最终显示仍然只有2个components。BayesianGaussianMixture ()针对权重分配默认包含两个,一个是包含狄利克雷分布的有限混合模型,另一个是包含狄利克雷过程的无限混合模型。
此处关于“狄利克雷分布”的详细介绍,可公众号“武汉AI算法研习”了解!
import itertools
import numpy as np
from scipy import linalg
import matplotlib.pyplot as plt
import matplotlib as mpl
from sklearn import mixture
color_iter = itertools.cycle(['navy', 'c', 'cornflowerblue', 'gold',
'darkorange'])
def plot_results(X, Y_, means, covariances, index, title):
splot = plt.subplot(2, 1, 1 + index)
for i, (mean, covar, color) in enumerate(zip(
means, covariances, color_iter)):
v, w = linalg.eigh(covar)
v = 2. * np.sqrt(2.) * np.sqrt(v)
u = w[0] / linalg.norm(w[0])
# as the DP will not use every component it has access to
# unless it needs it, we shouldn't plot the redundant
# components.
if not np.any(Y_ == i):
continue
plt.scatter(X[Y_ == i, 0], X[Y_ == i, 1], .8, color=color)
# Plot an ellipse to show the Gaussian component
angle = np.arctan(u[1] / u[0])
angle = 180. * angle / np.pi # convert to degrees
ell = mpl.patches.Ellipse(mean, v[0], v[1], 180. + angle, color=color)
ell.set_clip_box(splot.bbox)
ell.set_alpha(0.5)
splot.add_artist(ell)
plt.xlim(-9., 5.)
plt.ylim(-3., 6.)
plt.xticks(())
plt.yticks(())
plt.title(title)
# Number of samples per component
n_samples = 500
# Generate random sample, two components
np.random.seed(0)
C = np.array([[0., -0.1], [1.7, .4]])
X = np.r_[np.dot(np.random.randn(n_samples, 2), C),
.7 * np.random.randn(n_samples, 2) + np.array([-6, 3])]
# Fit a Gaussian mixture with EM using five components
gmm = mixture.GaussianMixture(n_components=5, covariance_type='full').fit(X)
plot_results(X, gmm.predict(X), gmm.means_, gmm.covariances_, 0,
'Gaussian Mixture')
# Fit a Dirichlet process Gaussian mixture using five components
dpgmm = mixture.BayesianGaussianMixture(n_components=5,
covariance_type='full').fit(X)
plot_results(X, dpgmm.predict(X), dpgmm.means_, dpgmm.covariances_, 1,
'Bayesian Gaussian Mixture with a Dirichlet process prior')
plt.show()
【Gaussian Mixture Model Selection】
This example shows that model selection can be performed with Gaussian Mixture Models using information-theoretic criteria (BIC). Model selection concerns both the covariance type and the number of components in the model. In that case, AIC also provides the right result (not shown to save time), but BIC is better suited if the problem is to identify the right model. Unlike Bayesian procedures, such inferences are prior-free.
高斯混合模型中分量个数的选择决定了最终模型的优劣,利用BIC(贝叶斯信息准则)来选择高斯混合模型的分量数是常用方法,但如果利用变分贝叶斯混合BayesianGaussianMixture () 则可以成功避免高斯混合模型中分量数的选择。从最终的结果图可以看出选择2个分量模型和full的模型,所得BIC值最低。
关于BIC和AIC的更多简单介绍,可关注公众号“武汉AI算法研习”文章了解!
源码中GaussianMixture.bic(X)方法就是关于输入X的在当前模型下的BIC值。
import numpy as np
import itertools
from scipy import linalg
import matplotlib.pyplot as plt
import matplotlib as mpl
from sklearn import mixture
print(__doc__)
# Number of samples per component
n_samples = 500
# Generate random sample, two components
np.random.seed(0)
C = np.array([[0., -0.1], [1.7, .4]])
X = np.r_[np.dot(np.random.randn(n_samples, 2), C),
.7 * np.random.randn(n_samples, 2) + np.array([-6, 3])]
lowest_bic = np.infty
bic = []
n_components_range = range(1, 7)
cv_types = ['spherical', 'tied', 'diag', 'full']
for cv_type in cv_types:
for n_components in n_components_range:
# Fit a Gaussian mixture with EM
gmm = mixture.GaussianMixture(n_components=n_components,
covariance_type=cv_type)
gmm.fit(X)
bic.append(gmm.bic(X))
if bic[-1] < lowest_bic:
lowest_bic = bic[-1]
best_gmm = gmm
bic = np.array(bic)
color_iter = itertools.cycle(['navy', 'turquoise', 'cornflowerblue',
'darkorange'])
clf = best_gmm
bars = []
# Plot the BIC scores
plt.figure(figsize=(8, 6))
spl = plt.subplot(2, 1, 1)
for i, (cv_type, color) in enumerate(zip(cv_types, color_iter)):
xpos = np.array(n_components_range) + .2 * (i - 2)
bars.append(plt.bar(xpos, bic[i * len(n_components_range):
(i + 1) * len(n_components_range)],
width=.2, color=color))
plt.xticks(n_components_range)
plt.ylim([bic.min() * 1.01 - .01 * bic.max(), bic.max()])
plt.title('BIC score per model')
xpos = np.mod(bic.argmin(), len(n_components_range)) + .65 +\
.2 * np.floor(bic.argmin() / len(n_components_range))
plt.text(xpos, bic.min() * 0.97 + .03 * bic.max(), '*', fontsize=14)
spl.set_xlabel('Number of components')
spl.legend([b[0] for b in bars], cv_types)
# Plot the winner
splot = plt.subplot(2, 1, 2)
Y_ = clf.predict(X)
for i, (mean, cov, color) in enumerate(zip(clf.means_, clf.covariances_,
color_iter)):
v, w = linalg.eigh(cov)
if not np.any(Y_ == i):
continue
plt.scatter(X[Y_ == i, 0], X[Y_ == i, 1], .8, color=color)
# Plot an ellipse to show the Gaussian component
angle = np.arctan2(w[0][1], w[0][0])
angle = 180. * angle / np.pi # convert to degrees
v = 2. * np.sqrt(2.) * np.sqrt(v)
ell = mpl.patches.Ellipse(mean, v[0], v[1], 180. + angle, color=color)
ell.set_clip_box(splot.bbox)
ell.set_alpha(.5)
splot.add_artist(ell)
plt.xticks(())
plt.yticks(())
plt.title('Selected GMM: full model, 2 components')
plt.subplots_adjust(hspace=.35, bottom=.02)
plt.show()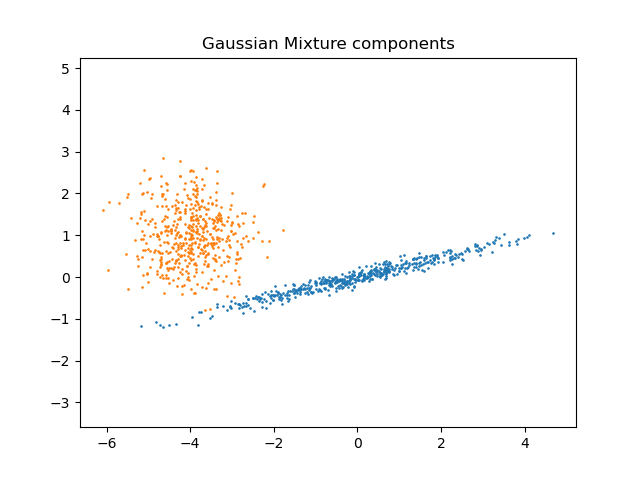
【GMM covariances】
We plot predicted labels on both training and held out test data using a variety of GMM covariance types on the iris dataset. We compare GMMs with spherical, diagonal, full, and tied covariance matrices in increasing order of performance. Although one would expect full covariance to perform best in general, it is prone to overfitting on small datasets and does not generalize well to held out test data.
GaussianMixture方法中自带了不同的选项来约束不同估类的协方差:spherical,diagonal,tied 或 full 协方差。
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets
from sklearn.mixture import GaussianMixture
from sklearn.model_selection import StratifiedKFold
print(__doc__)
colors = ['navy', 'turquoise', 'darkorange']
def make_ellipses(gmm, ax):
for n, color in enumerate(colors):
if gmm.covariance_type == 'full':
covariances = gmm.covariances_[n][:2, :2]
elif gmm.covariance_type == 'tied':
covariances = gmm.covariances_[:2, :2]
elif gmm.covariance_type == 'diag':
covariances = np.diag(gmm.covariances_[n][:2])
elif gmm.covariance_type == 'spherical':
covariances = np.eye(gmm.means_.shape[1]) * gmm.covariances_[n]
v, w = np.linalg.eigh(covariances)
u = w[0] / np.linalg.norm(w[0])
angle = np.arctan2(u[1], u[0])
angle = 180 * angle / np.pi # convert to degrees
v = 2. * np.sqrt(2.) * np.sqrt(v)
ell = mpl.patches.Ellipse(gmm.means_[n, :2], v[0], v[1],
180 + angle, color=color)
ell.set_clip_box(ax.bbox)
ell.set_alpha(0.5)
ax.add_artist(ell)
ax.set_aspect('equal', 'datalim')
iris = datasets.load_iris()
# Break up the dataset into non-overlapping training (75%) and testing
# (25%) sets.
skf = StratifiedKFold(n_splits=4)
# Only take the first fold.
train_index, test_index = next(iter(skf.split(iris.data, iris.target)))
X_train = iris.data[train_index]
y_train = iris.target[train_index]
X_test = iris.data[test_index]
y_test = iris.target[test_index]
n_classes = len(np.unique(y_train))
# Try GMMs using different types of covariances.
estimators = {cov_type: GaussianMixture(n_components=n_classes,
covariance_type=cov_type, max_iter=20, random_state=0)
for cov_type in ['spherical', 'diag', 'tied', 'full']}
n_estimators = len(estimators)
plt.figure(figsize=(3 * n_estimators // 2, 6))
plt.subplots_adjust(bottom=.01, top=0.95, hspace=.15, wspace=.05,
left=.01, right=.99)
for index, (name, estimator) in enumerate(estimators.items()):
# Since we have class labels for the training data, we can
# initialize the GMM parameters in a supervised manner.
estimator.means_init = np.array([X_train[y_train == i].mean(axis=0)
for i in range(n_classes)])
# Train the other parameters using the EM algorithm.
estimator.fit(X_train)
h = plt.subplot(2, n_estimators // 2, index + 1)
make_ellipses(estimator, h)
for n, color in enumerate(colors):
data = iris.data[iris.target == n]
plt.scatter(data[:, 0], data[:, 1], s=0.8, color=color,
label=iris.target_names[n])
# Plot the test data with crosses
for n, color in enumerate(colors):
data = X_test[y_test == n]
plt.scatter(data[:, 0], data[:, 1], marker='x', color=color)
y_train_pred = estimator.predict(X_train)
train_accuracy = np.mean(y_train_pred.ravel() == y_train.ravel()) * 100
plt.text(0.05, 0.9, 'Train accuracy: %.1f' % train_accuracy,
transform=h.transAxes)
y_test_pred = estimator.predict(X_test)
test_accuracy = np.mean(y_test_pred.ravel() == y_test.ravel()) * 100
plt.text(0.05, 0.8, 'Test accuracy: %.1f' % test_accuracy,
transform=h.transAxes)
plt.xticks(())
plt.yticks(())
plt.title(name)
plt.legend(scatterpoints=1, loc='lower right', prop=dict(size=12))
plt.show()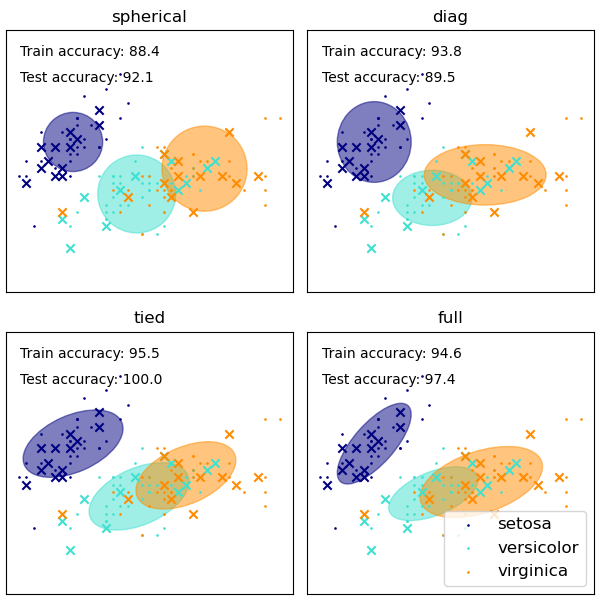
【Gaussian Mixture Model Sine Curve】
This example demonstrates the behavior of Gaussian mixture models fit on data that was not sampled from a mixture of Gaussian random variables. The dataset is formed by 100 points loosely spaced following a noisy sine curve. There is therefore no ground truth value for the number of Gaussian components.
在下图中,我们将拟合一个并不能被高斯混合模型很好描述的数据集。 调整 BayesianGaussianMixture类的参数 weight_concentration_prior,这个参数决定了用来拟合数据的分量数。我们在最后两个图上展示了从两个混合结果产生的随机抽样。
import itertools
import numpy as np
from scipy import linalg
import matplotlib.pyplot as plt
import matplotlib as mpl
from sklearn import mixture
print(__doc__)
color_iter = itertools.cycle(['navy', 'c', 'cornflowerblue', 'gold',
'darkorange'])
def plot_results(X, Y, means, covariances, index, title):
splot = plt.subplot(5, 1, 1 + index)
for i, (mean, covar, color) in enumerate(zip(
means, covariances, color_iter)):
v, w = linalg.eigh(covar)
v = 2. * np.sqrt(2.) * np.sqrt(v)
u = w[0] / linalg.norm(w[0])
# as the DP will not use every component it has access to
# unless it needs it, we shouldn't plot the redundant
# components.
if not np.any(Y == i):
continue
plt.scatter(X[Y == i, 0], X[Y == i, 1], .8, color=color)
# Plot an ellipse to show the Gaussian component
angle = np.arctan(u[1] / u[0])
angle = 180. * angle / np.pi # convert to degrees
ell = mpl.patches.Ellipse(mean, v[0], v[1], 180. + angle, color=color)
ell.set_clip_box(splot.bbox)
ell.set_alpha(0.5)
splot.add_artist(ell)
plt.xlim(-6., 4. * np.pi - 6.)
plt.ylim(-5., 5.)
plt.title(title)
plt.xticks(())
plt.yticks(())
def plot_samples(X, Y, n_components, index, title):
plt.subplot(5, 1, 4 + index)
for i, color in zip(range(n_components), color_iter):
# as the DP will not use every component it has access to
# unless it needs it, we shouldn't plot the redundant
# components.
if not np.any(Y == i):
continue
plt.scatter(X[Y == i, 0], X[Y == i, 1], .8, color=color)
plt.xlim(-6., 4. * np.pi - 6.)
plt.ylim(-5., 5.)
plt.title(title)
plt.xticks(())
plt.yticks(())
# Parameters
n_samples = 100
# Generate random sample following a sine curve
np.random.seed(0)
X = np.zeros((n_samples, 2))
step = 4. * np.pi / n_samples
for i in range(X.shape[0]):
x = i * step - 6.
X[i, 0] = x + np.random.normal(0, 0.1)
X[i, 1] = 3. * (np.sin(x) + np.random.normal(0, .2))
plt.figure(figsize=(10, 10))
plt.subplots_adjust(bottom=.04, top=0.95, hspace=.2, wspace=.05,
left=.03, right=.97)
# Fit a Gaussian mixture with EM using ten components
gmm = mixture.GaussianMixture(n_components=10, covariance_type='full',
max_iter=100).fit(X)
plot_results(X, gmm.predict(X), gmm.means_, gmm.covariances_, 0,
'Expectation-maximization')
dpgmm = mixture.BayesianGaussianMixture(
n_components=10, covariance_type='full', weight_concentration_prior=1e-2,
weight_concentration_prior_type='dirichlet_process',
mean_precision_prior=1e-2, covariance_prior=1e0 * np.eye(2),
init_params="random", max_iter=100, random_state=2).fit(X)
plot_results(X, dpgmm.predict(X), dpgmm.means_, dpgmm.covariances_, 1,
"Bayesian Gaussian mixture models with a Dirichlet process prior "
r"for $\gamma_0=0.01$.")
X_s, y_s = dpgmm.sample(n_samples=2000)
plot_samples(X_s, y_s, dpgmm.n_components, 0,
"Gaussian mixture with a Dirichlet process prior "
r"for $\gamma_0=0.01$ sampled with $2000$ samples.")
dpgmm = mixture.BayesianGaussianMixture(
n_components=10, covariance_type='full', weight_concentration_prior=1e+2,
weight_concentration_prior_type='dirichlet_process',
mean_precision_prior=1e-2, covariance_prior=1e0 * np.eye(2),
init_params="kmeans", max_iter=100, random_state=2).fit(X)
plot_results(X, dpgmm.predict(X), dpgmm.means_, dpgmm.covariances_, 2,
"Bayesian Gaussian mixture models with a Dirichlet process prior "
r"for $\gamma_0=100$")
X_s, y_s = dpgmm.sample(n_samples=2000)
plot_samples(X_s, y_s, dpgmm.n_components, 1,
"Gaussian mixture with a Dirichlet process prior "
r"for $\gamma_0=100$ sampled with $2000$ samples.")
plt.show()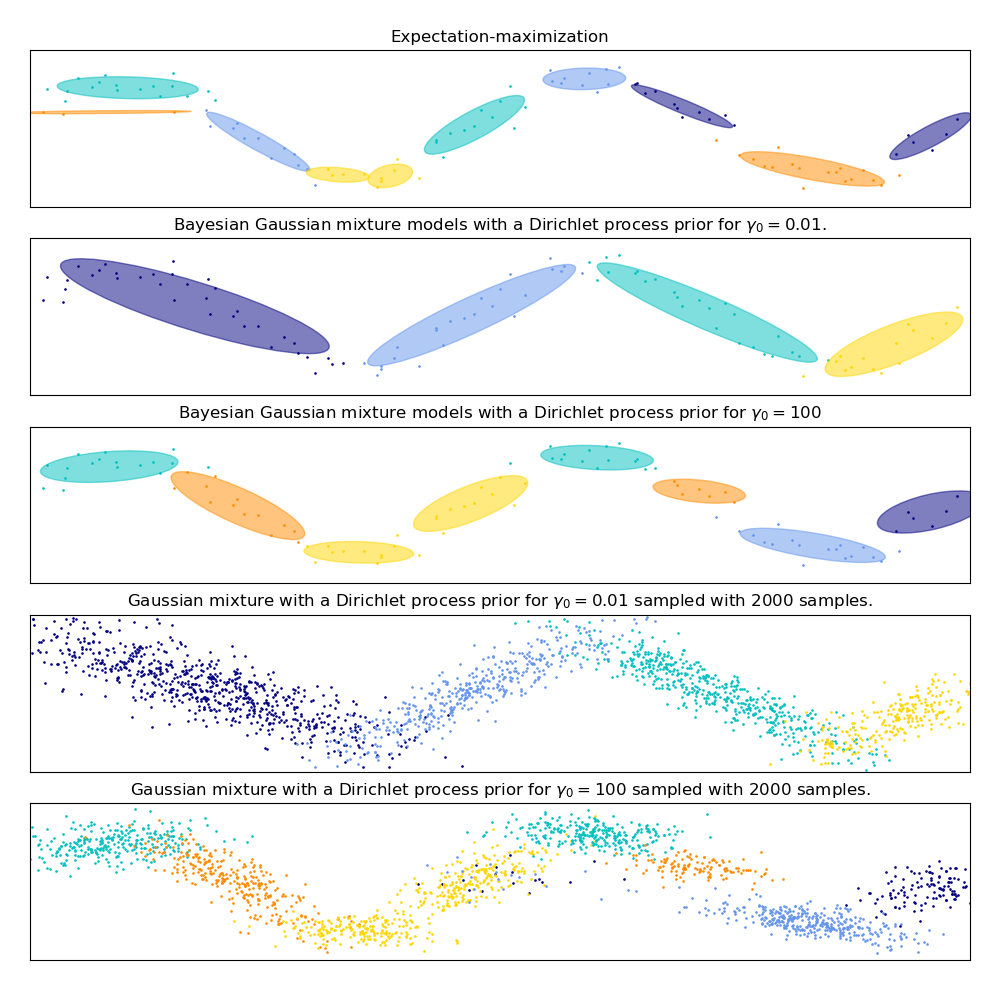
【Concentration Prior Type Analysis of Variation Bayesian Gaussian Mixture】
This example plots the ellipsoids obtained from a toy dataset (mixture of three Gaussians) fitted by the BayesianGaussianMixture class models with a Dirichlet distribution prior (weight_concentration_prior_type='dirichlet_distribution') and a Dirichlet process prior (weight_concentration_prior_type='dirichlet_process'). On each figure, we plot the results for three different values of the weight concentration prior.
下图比较了不同类型的权重浓度先验(参数 weight_concentration_prior_type) 不同的 weight_concentration_prior的值获得的结果。 在这里,我们可以发现 weight_concentration_prior参数的值对获得的有效的激活分量数(即权重较大的分量的数量)有很大影响。 我们也能注意到当先验是 ‘dirichlet_distribution’ 类型时,大的浓度权重先验会导致更均匀的权重,然而 ‘dirichlet_process’ 类型(默认类型)却不是这样。
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from sklearn.mixture import BayesianGaussianMixture
print(__doc__)
def plot_ellipses(ax, weights, means, covars):
for n in range(means.shape[0]):
eig_vals, eig_vecs = np.linalg.eigh(covars[n])
unit_eig_vec = eig_vecs[0] / np.linalg.norm(eig_vecs[0])
angle = np.arctan2(unit_eig_vec[1], unit_eig_vec[0])
# Ellipse needs degrees
angle = 180 * angle / np.pi
# eigenvector normalization
eig_vals = 2 * np.sqrt(2) * np.sqrt(eig_vals)
ell = mpl.patches.Ellipse(means[n], eig_vals[0], eig_vals[1],
180 + angle, edgecolor='black')
ell.set_clip_box(ax.bbox)
ell.set_alpha(weights[n])
ell.set_facecolor('#56B4E9')
ax.add_artist(ell)
def plot_results(ax1, ax2, estimator, X, y, title, plot_title=False):
ax1.set_title(title)
ax1.scatter(X[:, 0], X[:, 1], s=5, marker='o', color=colors[y], alpha=0.8)
ax1.set_xlim(-2., 2.)
ax1.set_ylim(-3., 3.)
ax1.set_xticks(())
ax1.set_yticks(())
plot_ellipses(ax1, estimator.weights_, estimator.means_,
estimator.covariances_)
ax2.get_xaxis().set_tick_params(direction='out')
ax2.yaxis.grid(True, alpha=0.7)
for k, w in enumerate(estimator.weights_):
ax2.bar(k, w, width=0.9, color='#56B4E9', zorder=3,
align='center', edgecolor='black')
ax2.text(k, w + 0.007, "%.1f%%" % (w * 100.),
horizontalalignment='center')
ax2.set_xlim(-.6, 2 * n_components - .4)
ax2.set_ylim(0., 1.1)
ax2.tick_params(axis='y', which='both', left=False,
right=False, labelleft=False)
ax2.tick_params(axis='x', which='both', top=False)
if plot_title:
ax1.set_ylabel('Estimated Mixtures')
ax2.set_ylabel('Weight of each component')
# Parameters of the dataset
random_state, n_components, n_features = 2, 3, 2
colors = np.array(['#0072B2', '#F0E442', '#D55E00'])
covars = np.array([[[.7, .0], [.0, .1]],
[[.5, .0], [.0, .1]],
[[.5, .0], [.0, .1]]])
samples = np.array([200, 500, 200])
means = np.array([[.0, -.70],
[.0, .0],
[.0, .70]])
# mean_precision_prior= 0.8 to minimize the influence of the prior
estimators = [
("Finite mixture with a Dirichlet distribution\nprior and "
r"$\gamma_0=$", BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_distribution",
n_components=2 * n_components, reg_covar=0, init_params='random',
max_iter=1500, mean_precision_prior=.8,
random_state=random_state), [0.001, 1, 1000]),
("Infinite mixture with a Dirichlet process\n prior and" r"$\gamma_0=$",
BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_process",
n_components=2 * n_components, reg_covar=0, init_params='random',
max_iter=1500, mean_precision_prior=.8,
random_state=random_state), [1, 1000, 100000])]
# Generate data
rng = np.random.RandomState(random_state)
X = np.vstack([
rng.multivariate_normal(means[j], covars[j], samples[j])
for j in range(n_components)])
y = np.concatenate([np.full(samples[j], j, dtype=int)
for j in range(n_components)])
# Plot results in two different figures
for (title, estimator, concentrations_prior) in estimators:
plt.figure(figsize=(4.7 * 3, 8))
plt.subplots_adjust(bottom=.04, top=0.90, hspace=.05, wspace=.05,
left=.03, right=.99)
gs = gridspec.GridSpec(3, len(concentrations_prior))
for k, concentration in enumerate(concentrations_prior):
estimator.weight_concentration_prior = concentration
estimator.fit(X)
plot_results(plt.subplot(gs[0:2, k]), plt.subplot(gs[2, k]), estimator,
X, y, r"%s$%.1e$" % (title, concentration),
plot_title=k == 0)
plt.show()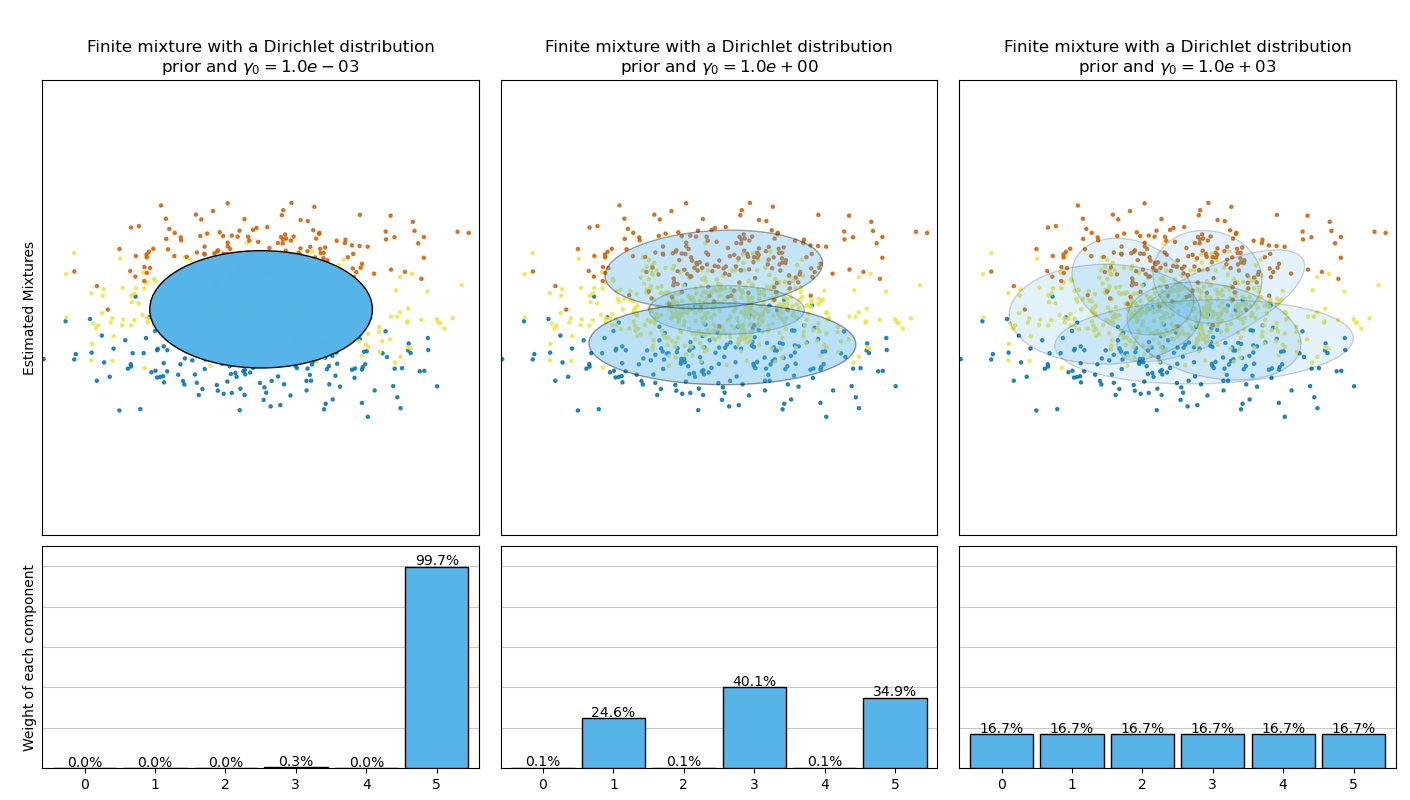
GMM模型中的超参数convariance_type控制这每个簇的形状自由度。
它的默认设置是convariance_type=’diag’,意思是簇在每个维度的尺寸都可以单独设置,但椭圆边界的主轴要与坐标轴平行。
covariance_type=’spherical’时模型通过约束簇的形状,让所有维度相等。这样得到的聚类结果和k-means聚类的特征是相似的,虽然两者并不完全相同。
covariance_type=’full’时,该模型允许每个簇在任意方向上用椭圆建模。
作为一种生成模型,GMM提供了一种确定数据集最优成分数量的方法。由于生成模型本身就是数据集的概率分布,因此可以利用模型来评估数据的似然估计,并利用交叉检验防止过拟合。Scikit-Learn的GMM评估器内置了两种纠正过拟合的标准分析方法:赤池信息量准则(AIC)和贝叶斯信息准则(BIC)






















 2936
2936











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








