Like all infectious diseases, Covid-19 transmission happens on a very local level. It does not just jump randomly around the world, it has to pass from person to person, and these people will have to travel to a new place to spread the virus there. It is for this reason that networks have often been used to help model the spread of diseases, whether it’s by using a social network to model the spread of the disease between people, or geographic networks to help us predict outbreaks based on what’s going on in neighbouring places.
像所有传染病一样,Covid-19传播发生在非常局部的水平。 它不仅在世界范围内随机跳跃,还必须在人与人之间传递,而且这些人将不得不前往新的地方以在那里传播病毒。 因此,网络经常被用来帮助对疾病的传播进行建模,无论是通过使用社交网络对人与人之间疾病的传播进行建模,还是通过地理网络来帮助我们根据疫情进行预测邻近的地方 。
In this blog series, we will be focusing on geographic networks, where the nodes are places, and the edges link two places if they neighbour each other. Ultimately we will try and use such a network to predict Covid cases in one area based on Covid cases in neighbouring areas.
在本博客系列中,我们将重点关注地理网络,其中节点是地点,并且如果边彼此相邻,则边将链接两个地点。 最终,我们将尝试并使用这样的网络,根据邻近地区的Covid病例预测一个地区的Covid病例。
The accuracy of this approach will be limited as it does not take account of epidemiological factors (such as herd immunity and social distancing) or other ways areas are linked (such as trains). However, this blog series still provides a helpful guide for visualising networks and using them to create features for predictive analysis. It also shows that network analysis can be a helpful piece of a larger Covid model.
这种方法的准确性将受到限制,因为它没有考虑到流行病学因素(例如畜群免疫力和社会距离)或其他地区之间的联系方式(例如火车)。 但是,该博客系列仍然为可视化网络以及使用网络创建用于预测分析的功能提供了有用的指南。 它还表明,网络分析可能是较大的Covid模型的有用部分。
In the UK, the smallest areas of geography that Covid cases are published on are called Middle Layer Super Output Areas (MSOAs) so we will focus on these. You can see this data in an interactive map here, and download it here.
在英国,Covid案例发布的最小地理区域称为中间层超级输出区域(MSOA),因此我们将重点关注这些区域。 您可以在此处的交互式地图中查看此数据,然后在此处下载。
In this first part, we will create some visualisations to motivate why such an approach might work, then we will create the network seen in the above image. We will showcase code as we go along, and also, all of the code can be found on github.
在第一部分中,我们将创建一些可视化效果来激发为什么这种方法可行的原因,然后我们将创建上图所示的网络。 我们将继续展示代码,并且所有代码都可以在github上找到。
These are all the packages that were used to create the visualisations shown in this post.
这些都是用于创建本文中显示的可视化效果的所有程序包。
# Set up ----
# Load packages
library(dplyr)
library(readr) # Loading the data
library(tidyr)
library(sf) # For the maps
library(sp) # Transform coordinates
library(ggplot2)
library(viridis)
library(gganimate) # For the animated map# Network packages
library(igraph) # build network
library(spdep) # builds network
library(tidygraph)
library(ggraph) # for plotting networks动机 (Motivation)
In order to see why this approach might work we will take a look at Leicester, which had a spike in Covid cases in June leading to the UKs first local lockdown. We are going to visualise the spread of Covid in the area by creating maps using the {ggplot2} and {sf} package, and animate these using the {gganimate} package.
为了弄清楚这种方法为何行得通,我们将看一下莱斯特,莱斯特在6月的Covid案件中激增,导致英国首次本地封锁。 我们将使用{ggplot2}和{sf}包创建地图,并使用{gganimate}包为其设置动画,以可视化方式展示Covid在该地区的分布。
We need to download the case data (which we can do directly from the URL), and get it into the right format. The initial table is wide (with one row per MSOA, and one column per week) and some of the weeks have no data, so we take advantage of the relatively new {tidyr} and {dplyr} functions where, pivot_longer and across to clean this data.
我们需要下载案例数据(可以直接从URL执行),并将其转换为正确的格式。 最初的表是宽(每MSOA一行,每周一列)和一些周没有数据,所以我们利用相对较新的{tidyr}和{dplyr}功能where , pivot_longer和across到干净这个数据。
n.b. Since publishing this story, the last_7_days column in the raw data has been renamed to latest_7_days.
自发布此故事以来,原始数据中的last_7_days列已重命名为latest_7_days 。
url <- 'https://c19downloads.azureedge.net/downloads/msoa_data/MSOAs_latest.csv'
msoa <- readr::read_csv(url, col_types = cols()) %>%
# Drop columns that are all NA
select(where(~!all(is.na(.)))) %>%
# Pivot longer
pivot_longer(dplyr::starts_with("wk_"),
names_to = "week",
values_to = "cases",
names_prefix = "wk_") %>%
# Turn week to numeric
mutate(week = as.integer(week)) %>%
# Turn NAs into 0s
mutate(across(c(last_7_days, cases),
.fns = ~ifelse(is.na(.), 0, .)))The resulting table looks like this (I cut the first few columns out to simplify it). The first two columns tell us the Local Authority District (LAD) which is a larger area than MSOA. The next two columns tell us the MSOA. The final two columns tell us the week, and how many positive cases there were in that week. The “last_7_days” column tells us the number of positive cases between the last full week and today.
结果表如下所示(为简化起见,我删掉了前几列)。 前两列告诉我们地方当局区(LAD),其面积比MSOA大。 接下来的两列告诉我们MSOA。 最后两列告诉我们一周以及该周有多少积极病例。 “ last_7_days”列告诉我们最近一整周到今天之间的阳性病例数。

The first thing we should do is check that the Leicester spike really does appear in the data, which we can do by plotting a simple line graph.
我们应该做的第一件事是检查莱斯特峰是否确实出现在数据中,这可以通过绘制简单的线图来完成。
msoa %>%
group_by(lad19_nm, week) %>%
summarise(cases = sum(cases))%>%
filter(lad19_nm == "Leicester") %>%
ggplot(aes(x = week, y = cases)) +
geom_line(size = 1) +
theme_minimal() +
ggtitle("Time series gaph of cases for Leicester")
We see that there is a large spike in the data, peaking at 500 cases in week 25.
我们看到数据有一个很大的峰值,在第25周达到500例峰值。
The next step is to see how this is distributed amongst the MSOAs. We could do this as another line graph (and in fact, we do in the github code) but that won’t give us the same insight as if we plot it on a map.
下一步是查看如何在MSOA之间进行分配。 我们可以将其作为另一个折线图来完成(实际上,我们在github代码中完成),但这不会像在地图上绘制图一样给我们提供相同的见解。
To plot a map, we have to download the shape file of MSOAs in the UK, which you can do here. After saving and unzipping the whole folder, we can load it in the following way:
要绘制地图,我们必须在英国下载MSOA的形状文件,您可以在此处进行操作 。 保存并解压缩整个文件夹后,我们可以通过以下方式加载它:
msoa_sf <- sf::st_read("data/maps/Middle_Layer_Super_Output_Areas__December_2011__Boundaries_EW_BFE.shp")Plotting the MSOAs of Leicester is as simple as restricting the shape file down to just those that are in Leicester (we do this by inner joining), and then adding a geom_sf() to a ggplot. To plot the case data we restrict to a particular week (let’s choose the peak of week 25) and join on the cases data too. We can now fill the MSOA areas based on how many positive cases they had in week 25.
绘制莱斯特的MSOA就像将形状文件限制为仅位于莱斯特中的形状文件一样简单(我们通过内部geom_sf()来完成),然后将geom_sf()添加到ggplot中。 为了绘制案例数据,我们限制在特定的一周(让我们选择第25周的峰值),并加入案例数据。 现在,我们可以根据他们在第25周有多少阳性病例来填充MSOA区域。
msoa_data_wk25 <- msoa %>%
filter(lad19_nm == "Leicester",
week == 25)# We see that covid cases are clustered in several MSOAs
msoa_sf %>%
inner_join(msoa_data_wk25 %>% distinct(msoa11_cd, cases),
by = c("MSOA11CD" = "msoa11_cd")) %>%
ggplot()+
geom_sf(aes(geometry = geometry, fill = cases)) +
# Everything past this point is just formatting:
scale_fill_viridis_c() +
theme_void() +
ggtitle(label = "MSOA map for Leicester",
subtitle = "The colour shows Covid cases during week 25")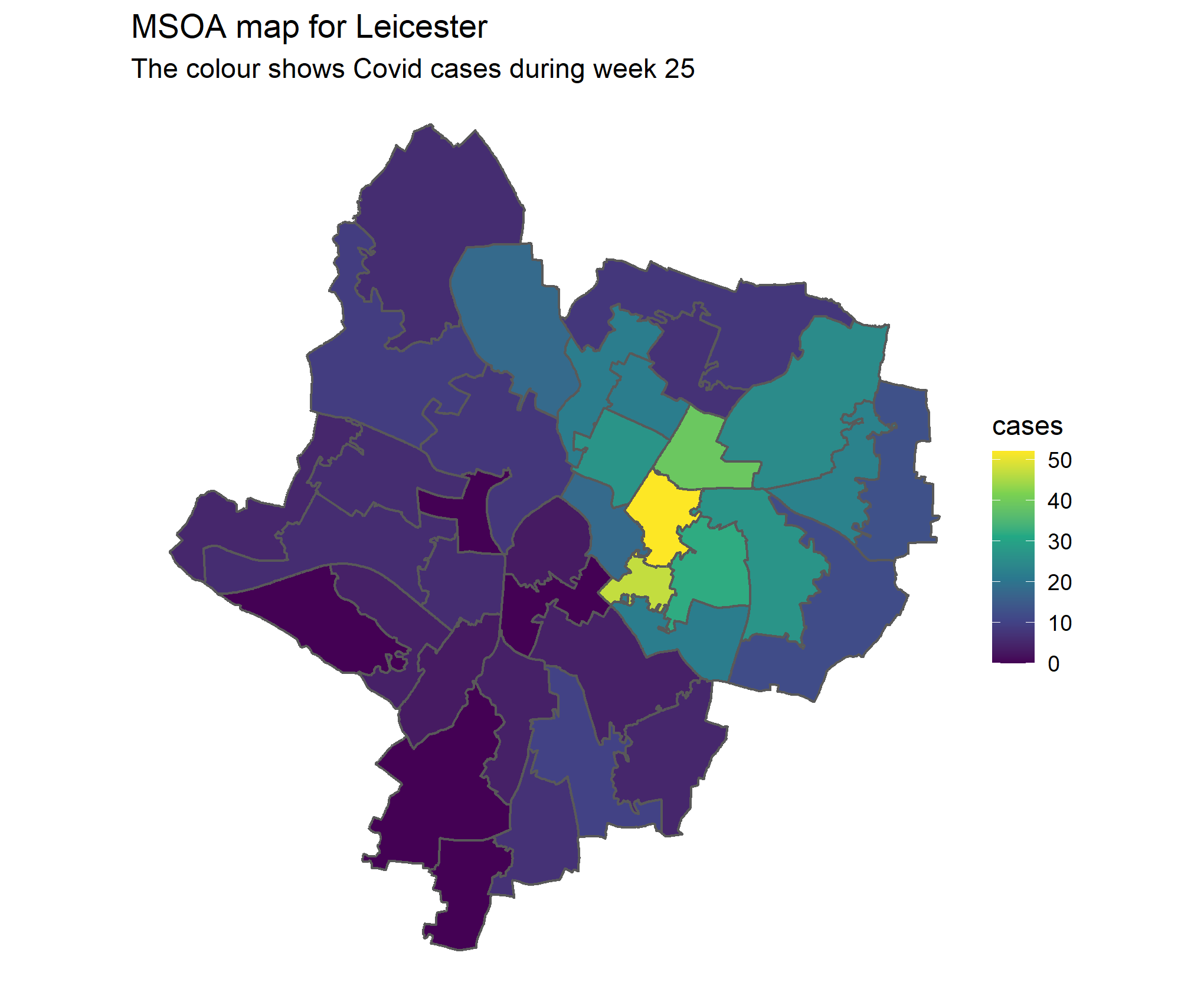
This is where we get the first indication that networks will be useful. We see that Neighbouring MSOAs have cases similar to neighbouring areas. This makes sense. If you have an outbreak in one place you expect it to spread to neighbouring places.
这是我们第一个表明网络将有用的迹象。 我们看到,邻近MSOA的情况与邻近地区相似。 这很有道理。 如果您在某个地方爆发了疫情,您希望它会蔓延到附近的地方。
The one downside of visualising data like this, is it’s hard to see how the cases change over time. That’s where {gganimate} comes in. {gganimate} is a great package that let’s you animate your graphs based on a particular variable. It allows us to create the above map for every week, and then combines it together into a gif for us.
像这样可视化数据的一个缺点是,很难看到案例随着时间的变化。 这就是{gganimate}的来源。{gganimate}是一个很棒的程序包,可让您基于特定变量对图形进行动画处理。 它使我们能够每周创建上面的地图,然后将其组合在一起成为gif。
# gganimate
# Need to install transformr for this to work
p <- msoa_sf %>%
inner_join(msoa %>% filter(lad19_nm == "Leicester"),
by = c("MSOA11CD" = "msoa11_cd")) %>%
ggplot(aes(group = week)) +
geom_sf(aes(fill = cases)) +
scale_fill_viridis_c() +
transition_time(week) +
labs(title = paste0("New covid cases for MSOAs within Leicester"),
subtitle = "Week = {frame_time}") +
theme_void()# We define this to help us pick the right number of frames
num_weeks <- n_distinct(msoa$week)animate(p, nframes = num_weeks, fps = 1, end_pause = 4)We see that up to transition_time() , this code looks really similar to our last graph. The only difference is we don’t filter the data to a specific week, and we include a group = week in the aes() for the plot. transition_time() is how we instruct it to use week as the variable to animate over.
我们看到,直到transition_time()为止,此代码看起来与我们的上一张图非常相似。 唯一的区别是我们不将数据过滤到特定的星期,而是在绘图的aes()包括group = week 。 transition_time()是我们指示它使用week作为变量进行动画处理的方式。
The final bit of code just helps us specify a good number of frames so each week gets the same number. The result is below.
最后的代码只是帮助我们指定了很多帧,因此每周获得的帧数都是相同的。 结果如下。
Again, the importance of neighbouring areas shines through. High case numbers spread from one area to neighbouring areas. Also, just before the peak in numbers for a single MSOA (which happens in week 27) we see the neighbouring areas all have high case numbers too. Would network analysis have allowed us to predict that this one particular area would have such a spike in cases in week 27? There’s only one way to find out.
再次,邻近地区的重要性得以体现。 高病例数从一个区域扩展到相邻区域。 另外,就在单个MSOA数量达到峰值之前(发生在第27周),我们看到邻近地区的案例数量也很高。 网络分析是否可以让我们预测,在第27周的情况下,这一特定区域会出现这种峰值? 只有一种方法可以找出答案。
网络 (The Network)
Now we know why networks can help us, let’s actually build one. To define a network, it is enough to know what all the nodes are, and how they join together (i.e. the edges).
现在我们知道了网络为什么可以帮助我们,让我们实际构建一个。 要定义一个网络,只需知道所有节点是什么,以及它们如何连接在一起(即边缘)就足够了。
In our case the nodes are simple, these are just the MSOAs. We know we want the edges to be such that two nodes are joined if the two MSOAs they represent are neighbours on our map. Fortunately, the poly2nb() function (read “polygon to neighbours”) from the {spdep} package helps us do just this. It can take a shape file, and tell us which areas are neighbours.
在我们的例子中,节点很简单,它们只是MSOA。 我们知道,如果它们表示的两个MSOA在我们的地图上是相邻的,则我们希望边成为两个节点相连。 幸运的是,来自{spdep}包的poly2nb()函数(读取为“向邻方多边形”)可以帮助我们做到这一点。 它可以提取形状文件,并告诉我们哪些区域是邻居。
# First we just need to restrict to Leicester MSOAs for one week.
leicester_sf <- msoa_sf %>%
inner_join(msoa %>% filter(lad19_nm == "Leicester", week == 27) ,
by = c("MSOA11CD" = "msoa11_cd"))# Use the poly2nb to get the neighbourhood of each area
leicester_msoa_neighbours <- spdep::poly2nb(leicester_sf)We can then use the nb2mat() function (read “neighbours to matrix”) to turn this into an adjacency matrix. For a network, an adjacency matrix is such that cell i, j is equal to 1 if there is an edge connecting node i to node j, and a 0 otherwise. Hence an adjacency matrix defines a network, and in fact, we can use the graph_from_adjacency_matrix function from the {igraph} package to create a network object.
然后,我们可以使用nb2mat()函数(阅读“矩阵邻居”)将其转换为邻接矩阵。 对于网络,邻接矩阵是这样的:如果存在将节点i连接到节点j的边缘,则单元i,j等于1 ,否则为0。 因此,邻接矩阵定义了一个网络,实际上,我们可以使用{igraph}包中的graph_from_adjacency_matrix函数来创建网络对象。
# Use nb2mat to turn this into an adjacency matrix
adj_mat <- spdep::nb2mat(leicester_msoa_neighbours, style = "B")
rownames(adj_mat) <- leicester_sf$msoa11_hclnm
colnames(adj_mat) <- leicester_sf$msoa11_hclnm# Use graph_from_adjacency_matrix to create a graph object from the adjacency matrix
leicester_network <- igraph::graph_from_adjacency_matrix(adj_mat, mode = "undirected")You could stop here, but I have found that the {tidygraph} package provides an amazing way of manipulating network objects using standard {dplyr} verbs (which we will see later). So we turn this network object into a tidy graph object using as_tbl_graph().
您可以在这里停下来,但是我发现{tidygraph}包提供了一种使用标准{dplyr}动词来操作网络对象的惊人方法(我们将在后面介绍)。 因此,我们使用as_tbl_graph()将网络对象转换为整洁的图形对象。
# Use as_tbl_graph to turn this into a tidygraph
leicester_network <- igraph::graph_from_adjacency_matrix(adj_mat, mode = "undirected") %>%
tidygraph::as_tbl_graph()Now we have created this object, how do we interact with it?
现在我们已经创建了这个对象,我们如何与它交互?
Printing the object shows our network has 37 nodes and 91 edges. We also see that it is defined by two tibbles, one to define the node data, and one to define the edge data.
打印对象表明我们的网络具有37个节点和91个边缘。 我们还看到它由两个小节定义,一个小节定义节点数据,另一个小节定义边缘数据。
> leicester_network
# A tbl_graph: 37 nodes and 91 edges
#
# An undirected simple graph with 1 component
#
# Node Data: 37 x 1 (active)
name
<chr>
1 Beamont Park
2 Rushey Mead North
3 Stocking Farm & Mowmacre
4 Bradgate Heights & Beaumont Leys
5 Rushey Mead South
6 Belgrave North West
# ... with 31 more rows
#
# Edge Data: 91 x 2
from to
<int> <int>
1 1 3
2 1 4
3 2 3
# ... with 88 more rowsNote that the node data just contains the names of the MSOA, and the edge data tells us which nodes are connected based on their position in the node data. For example, the first row of the edge data tells us “Beamont Park” and “Stocking Farm & Mowmacre” are neighbours.
请注意,节点数据仅包含MSOA的名称,边缘数据根据节点在节点数据中的位置告诉我们连接了哪些节点。 例如,边缘数据的第一行告诉我们“ Beamont Park”和“ Stocking Farm&Mowmacre”是邻居。
If we want to visualise the network, we can use {ggraph}, which is a network specific extension to {ggplot2} and works in much the same way. Here’s how we create a simple plot. The node positions are determined by an algorithm.
如果要可视化网络,可以使用{ggraph},它是{ggplot2}的网络特定扩展,其工作方式几乎相同。 这是我们创建简单图的方式。 节点位置由算法确定。
ggraph(leicester_network) +
geom_edge_link(colour = 'black', width = 2) +
geom_node_point(size = 5, colour = 'steelblue') +
theme_void() +
ggtitle("Network plot of MSOAs in Leicester",
subtitle = "Each node is an MSOA, and an edge joins two
nodes if they are neighbours")
It would be nice if we could overlay this network on top of our map from before. This would allow us to sense check the network, and make it easier to translate between the network and reality.
如果我们可以从以前将这个网络覆盖在地图顶部,那就太好了。 这将使我们能够感觉到对网络的检查,并使其更容易在网络与现实之间进行转换。
In order to achieve this, we first need to get central co-ordinates for each MSOA. Fortunately, the shape file we downloaded comes with a pair of latitude longitude coordinates for each MSOA. Unfortunately, the polygons themselves are under a different spatial projection. We use the following code to extract the co-ordinates and transform the projection.
为了实现这一目标,我们首先需要获取每个MSOA的中心坐标。 幸运的是,我们下载的形状文件为每个MSOA附带了一对纬度经度坐标。 不幸的是,多边形本身处于不同的空间投影下。 我们使用以下代码提取坐标并变换投影。
coords <- leicester_sf %>%
as.data.frame() %>%
select(LONG, LAT) %>%
sp::SpatialPoints(proj4string=CRS("+init=epsg:4326")) %>% # LAT LONG code
sp::spTransform(CRS("+init=epsg:27700")) %>% # UK grid code
as.data.frame() %>%
bind_cols(msoa11_hclnm = leicester_sf$msoa11_hclnm) # to allow us to bind oncoords looks like this:
coords看起来像这样:
LONG LAT msoa11_hclnm
<dbl> <dbl> <chr>
1 457176. 309094. Beamont Park
2 461975. 307767. Rushey Mead North
3 458317. 307856. Stocking Farm & Mowmacre
4 456550. 306747. Bradgate Heights & Beaumont Leys
5 461046. 307294. Rushey Mead South
6 459732. 307380. Belgrave North West
7 460299. 306733. Belgrave North East
8 458059. 305765. Abbey Park
9 462606. 306391. Hamilton & Humberstone
10 459885. 305741. Belgrave South
# ... with 27 more rowsBy binding on the MSOA names, it makes it really easy to join this onto our network object. We essentially want to left_join onto our nodes table. {tidygraph} makes this really easy. All we have to do is tell it we want to do something with the nodes table by using activate() then we can use the classic {dplyr} verbs we all know and love, like left_join().
通过绑定MSOA名称,可以很容易地将其加入到我们的网络对象中。 我们本质上是想left_join到我们的节点表上。 {tidygraph}使这变得非常容易。 我们要做的就是告诉我们我们想通过使用activate()对nodes表做一些事情,然后我们可以使用我们都知道并喜欢的经典{dplyr}动词,例如left_join() 。
# Join on cordinates and sf geometry
leicester_network <- leicester_network %>%
activate("nodes") %>%
left_join(coords, by = c("name" = "msoa11_hclnm"))At any time, you can use as_tibble() to extract the activated tibble (i.e. either nodes or edges). If we do that now, we see the co-ordinates have been successfully joined.
在任何时候,您都可以使用as_tibble()提取激活的小标题(即节点或边)。 如果现在执行此操作,则可以看到已成功加入坐标。
leicester_network %>%
as_tibble() %>%
head
name LONG LAT
<chr> <dbl> <dbl>
1 Beamont Park 457176. 309094.
2 Rushey Mead North 461975. 307767.
3 Stocking Farm & Mowmacre 458317. 307856.
4 Bradgate Heights & Beaumont Leys 456550. 306747.
5 Rushey Mead South 461046. 307294.
6 Belgrave North West 459732. 307380.Now we just create a ggraph plot, where we use the MSOA shape file for a geom_sf layer, and then overlay our network.
现在,我们仅创建一个ggraph图,在其中将MSOA形状文件用于geom_sf层,然后覆盖我们的网络。
ggraph(leicester_network, layout = "manual", x = LONG, y = LAT) +
geom_sf(data = leicester_sf, fill = "white") +
theme_void() +
geom_edge_link(colour = 'black', width = 2) +
geom_node_point(size = 5, colour = 'steelblue') +
ggtitle("MSOA network overlaying shapefile")
And just like that, we have shown the network is exactly what we wanted it to be; every node is an MSOA, and an edge connects two if they are neighbours.
就像这样,我们已经证明了网络正是我们想要的。 每个节点都是MSOA,如果它们是邻居,则边缘将连接两个。
下次 (Next time)
We have successfully shown why we might look to networks to help us understand how COVID spreads, and also how to build and visualise such a network.
我们已经成功地展示了为什么我们可能会寻求网络来帮助我们了解COVID的传播方式,以及如何构建和可视化这种网络。
Next time, we will see how the network structure allows us to create powerful features intuitively and quickly, and then use these new features to create predictive models.
下次,我们将看到网络结构如何使我们能够直观,快速地创建强大的功能,然后使用这些新功能来创建预测模型。
All the code used in this post can be found on GitHub where renv has been used to deal with package versions and dependencies.
翻译自: https://towardsdatascience.com/predicting-the-spread-of-covid-19-using-networks-in-r-2fee0013db91























 1530
1530

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








