单细胞生信分析教程
桓峰基因公众号推出单细胞生信分析教程并配有视频在线教程,目前整理出来的相关教程目录如下:
SCS【4】单细胞转录组数据可视化分析 (Seurat 4.0)
SCS【6】单细胞转录组之细胞类型自动注释 (SingleR)
SCS【7】单细胞转录组之轨迹分析 (Monocle 3) 聚类、分类和计数细胞
SCS【8】单细胞转录组之筛选标记基因 (Monocle 3)
SCS【9】单细胞转录组之构建细胞轨迹 (Monocle 3)
SCS【10】单细胞转录组之差异表达分析 (Monocle 3)
SCS【11】单细胞ATAC-seq 可视化分析 (Cicero)
SCS【12】单细胞转录组之评估不同单细胞亚群的分化潜能 (Cytotrace)
SCS【13】单细胞转录组之识别细胞对“基因集”的响应 (AUCell)
SCS【15】细胞交互:受体-配体及其相互作用的细胞通讯数据库 (CellPhoneDB)
SCS【16】从肿瘤单细胞RNA-Seq数据中推断拷贝数变化 (inferCNV)
SCS【17】从单细胞转录组推断肿瘤的CNV和亚克隆 (copyKAT)
SCS【18】细胞交互:受体-配体及其相互作用的细胞通讯数据库 (iTALK)
SCS【21】单细胞空间转录组可视化 (Seurat V5)
SCS【22】单细胞转录组之 RNA 速度估计 (Velocyto.R)
SCS【24】单细胞数据量化代谢的计算方法 (scMetabolism)
SCS【25】单细胞细胞间通信第一部分细胞通讯可视化(CellChat)
SCS【26】单细胞细胞间通信第二部分通信网络的系统分析(CellChat)
SCS【28】单细胞转录组加权基因共表达网络分析(hdWGCNA)
SCS【29】单细胞基因富集分析 (singleseqgset)
SCS【30】单细胞空间转录组学数据库(STOmics DB)
SCS【31】减少障碍,加速单细胞研究数据库(Single Cell PORTAL)
简 介
eQTL 研究对于理解基因组调控至关重要。遗传变异对基因调控的影响是细胞类型特异性和细胞环境相关的,因此在单细胞水平上研究 eQTL 是至关重要的。理想的解决方案是同时使用来自同一细胞的突变和表达数据。然而,目前这种单细胞配对数据的技术还不成熟。我们提出了一种新的方法,eQTLsingle 仅用 scRNA-seq 数据发现 eQTL,而不需要基因组数据。从 scRNA-seq 数据中检测突变,利用零膨胀负二项 (zero-inflated negative binomial, ZINB) 模型对不同基因型的基因表达进行建模,在单细胞水平上寻找基因型和表型之间的关联。基于胶质母细胞瘤和胶质瘤 scRNA-seq 数据集,eQTLsingle 发现了数百个细胞类型特异性肿瘤相关的 eQTL,其中大多数在批量eQTL研究中无法发现。对发现的 eQTL 的详细分析揭示了重要的潜在调控机制。eQTLsingle 是一个独特而强大的工具,可以利用大量的 scRNA-seq 资源进行单细胞 eQTL 研究。
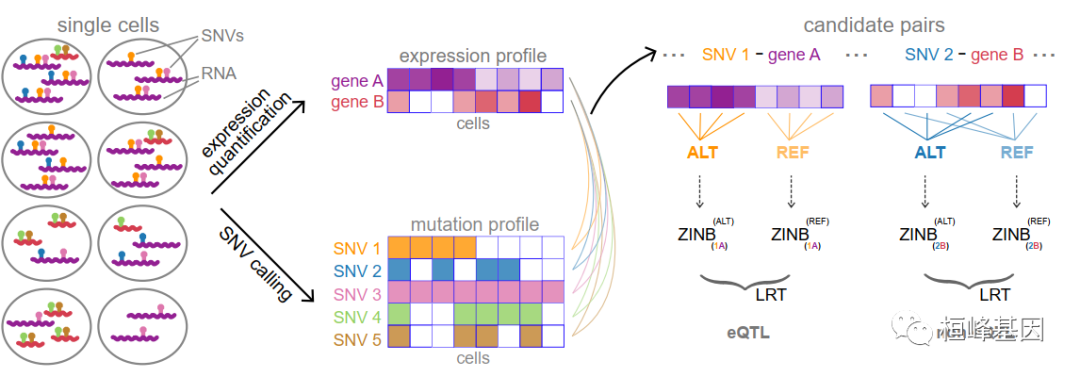
eQTLsingle has two modes. Cleaned-data mode: take input as SNV matrix and expression matrix from single cells. If you have had SNV and expression information of the same cell before, you can use this mode to discover eQTLs Raw-data mode: take input as raw scRNA-seq data (fastq data). eQTLsingle can generate both SNV matrix and gene matrix, and discovers eQTLs based them
软件包安装
if(!require(devtools))
install.packages("devtools")
devtools::install_github("horsedayday/eQTLsingle", build_vignettes = TRUE)Cleaned-data mode
eQTLsingle接受两个输入:snvMatrix和expressionMatrix。
snvMatrix:一个数据框,描述 SNV 的基因型(以细胞为单位的基因座),该数据框的行名是SNV id,该数据框的别名是CellId。(1) -1表示数据缺失(没有覆盖足够的读取);(2) 0表示该位点基因型为REF(非突变);(3) 1表示该基因座的基因型为ALT(突变);
expressionMatrix:一个数据框,描述基因表达(细胞的基因表达),该数据框的行名为Geneid,该数据框的别名为CellId。
Step1: Build SNV-gene pairs
只对有效的突变和基因进行eQTL分析。因此,需要首先选择高可靠性的SNV基因对(SNV和基因都覆盖足够的细胞)。
# Load eQTLsingle and test data (SNV matrix and expression matrix)
library(eQTLsingle)
library(knitr)## Warning: 程辑包'knitr'是用R版本4.2.3 来建造的data("toy_snvMatrix")
kable(toy_snvMatrix[1:5,])| cell_1 | cell_2 | cell_3 | cell_4 | cell_5 | cell_6 | cell_7 | cell_8 | cell_9 | cell_10 | cell_11 | cell_12 | cell_13 | cell_14 | cell_15 | cell_16 | cell_17 | cell_18 | cell_19 | cell_20 | cell_21 | cell_22 | cell_23 | cell_24 | cell_25 | cell_26 | cell_27 | cell_28 | cell_29 | cell_30 | cell_31 | cell_32 | cell_33 | cell_34 | cell_35 | cell_36 | cell_37 | cell_38 | cell_39 | cell_40 | cell_41 | cell_42 | cell_43 | cell_44 | cell_45 | cell_46 | cell_47 | cell_48 | cell_49 | cell_50 | cell_51 | cell_52 | cell_53 | cell_54 | cell_55 | cell_56 | cell_57 | cell_58 | cell_59 | cell_60 | cell_61 | cell_62 | cell_63 | cell_64 | cell_65 | cell_66 | cell_67 | cell_68 | cell_69 | cell_70 | cell_71 | cell_72 | cell_73 | cell_74 | cell_75 | cell_76 | cell_77 | cell_78 | cell_79 | cell_80 | cell_81 | cell_82 | cell_83 | cell_84 | cell_85 | cell_86 | cell_87 | cell_88 | cell_89 | cell_90 | cell_91 | cell_92 | cell_93 | cell_94 | cell_95 | cell_96 | cell_97 | cell_98 | cell_99 | cell_100 | cell_101 | cell_102 | cell_103 | cell_104 | cell_105 | cell_106 | cell_107 | cell_108 | cell_109 | cell_110 | cell_111 | cell_112 | cell_113 | cell_114 | cell_115 | cell_116 | cell_117 | cell_118 | cell_119 | cell_120 | cell_121 | cell_122 | cell_123 | cell_124 | cell_125 | cell_126 | cell_127 | cell_128 | cell_129 | cell_130 | cell_131 | cell_132 | cell_133 | cell_134 | cell_135 | cell_136 | cell_137 | cell_138 | cell_139 | cell_140 | cell_141 | cell_142 | cell_143 | cell_144 | cell_145 | cell_146 | cell_147 | cell_148 | cell_149 | cell_150 | cell_151 | cell_152 | cell_153 | cell_154 | cell_155 | cell_156 | cell_157 | cell_158 | cell_159 | cell_160 | cell_161 | cell_162 | cell_163 | cell_164 | cell_165 | cell_166 | cell_167 | cell_168 | cell_169 | cell_170 | cell_171 | cell_172 | cell_173 | cell_174 | cell_175 | cell_176 | cell_177 | cell_178 | cell_179 | cell_180 | cell_181 | cell_182 | cell_183 | cell_184 | cell_185 | cell_186 | cell_187 | cell_188 | cell_189 | cell_190 | cell_191 | cell_192 | cell_193 | cell_194 | cell_195 | cell_196 | cell_197 | cell_198 | cell_199 | cell_200 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNV_1 | 0 | 1 | 0 | 1 | 1 | -1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | -1 | 0 | 0 | 1 | 1 | 1 | -1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | -1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | -1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | -1 | 0 | -1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | -1 | 0 | 0 | -1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | -1 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | -1 | 0 | 0 | 0 | 0 | 1 | 1 | -1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | -1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | -1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | -1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 |
| SNV_2 | 1 | -1 | -1 | -1 | 0 | -1 | 1 | -1 | 1 | -1 | 1 | -1 | -1 | -1 | 1 | 0 | -1 | -1 | -1 | -1 | -1 | -1 | 0 | -1 | -1 | 1 | 1 | -1 | 0 | -1 | 0 | -1 | 0 | -1 | -1 | -1 | 1 | 0 | -1 | -1 | -1 | 0 | 0 | -1 | 1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | 0 | 1 | 0 | -1 | -1 | 1 | 0 | 1 | -1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | 1 | 1 | -1 | 0 | -1 | -1 | 0 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | -1 | 0 | -1 | 0 | -1 | 0 | -1 | -1 | -1 | -1 | 0 | -1 | 1 | 0 | -1 | -1 | -1 | -1 | 0 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 0 | -1 | -1 | 0 | -1 | 0 | -1 | -1 | -1 | 0 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 | 0 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 0 | -1 | 1 | 0 | -1 | 0 | -1 | -1 | 1 | 1 | -1 | -1 | 0 | -1 | 0 | 0 | 0 | 1 | -1 |
| SNV_3 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | -1 | 1 | 1 | -1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | -1 | 0 | 0 | 1 | -1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | -1 | 1 | -1 | -1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | -1 | 1 | 0 | 0 | -1 | 0 | -1 | 0 | 1 | -1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | -1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | -1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | -1 | 0 | 0 | -1 | 0 | 1 | 0 | -1 | 0 | 1 | 0 | 1 | 0 | 0 | -1 | 0 | -1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | -1 | -1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| SNV_4 | 0 | 1 | -1 | 1 | -1 | 1 | -1 | 0 | 0 | 1 | 0 | 0 | -1 | -1 | -1 | -1 | 0 | -1 | 0 | -1 | -1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 | -1 | -1 | 0 | -1 | -1 | 1 | -1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | 0 | 1 | -1 | -1 | -1 | -1 | 0 | -1 | -1 | -1 | -1 | -1 | 0 | 0 | -1 | -1 | -1 | 0 | -1 | -1 | 1 | 1 | -1 | -1 | -1 | -1 | -1 | 0 | -1 | -1 | -1 | 1 | -1 | 1 | -1 | -1 | 0 | 0 | -1 | 1 | -1 | -1 | -1 | 1 | 0 | 0 | -1 | -1 | -1 | 0 | -1 | -1 | -1 | -1 | -1 | 1 | 0 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 | -1 | -1 | 1 | -1 | 0 | -1 | 1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 1 | 1 | -1 | -1 | 1 | -1 | -1 | -1 | -1 | 1 | -1 | 1 | 0 | -1 | -1 | -1 | -1 | -1 | 1 | -1 | -1 | -1 | 1 | -1 | 1 | 1 | 1 | -1 | -1 | 0 | -1 | 1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | -1 | 0 | 1 | -1 | 0 | 1 | -1 | 1 | -1 | -1 | -1 | 0 | -1 | -1 | -1 | -1 | 0 | 0 | -1 | 1 | -1 | -1 | -1 | -1 | 0 | -1 | 1 | -1 |
| SNV_5 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | -1 | 1 | 1 | 0 | 0 | 0 | 1 | -1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | -1 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | -1 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | -1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | -1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | -1 | 0 | 0 | 0 | 1 | 1 | -1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | -1 | 0 | 0 | -1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 | -1 | -1 | 0 | 1 | 1 | -1 | 1 | 1 | -1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | -1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | -1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | -1 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | -1 | 0 | 1 | 0 | -1 | 0 |
data("toy_expressionMatrix")
kable(toy_expressionMatrix[1:5,])| cell_1 | cell_2 | cell_3 | cell_4 | cell_5 | cell_6 | cell_7 | cell_8 | cell_9 | cell_10 | cell_11 | cell_12 | cell_13 | cell_14 | cell_15 | cell_16 | cell_17 | cell_18 | cell_19 | cell_20 | cell_21 | cell_22 | cell_23 | cell_24 | cell_25 | cell_26 | cell_27 | cell_28 | cell_29 | cell_30 | cell_31 | cell_32 | cell_33 | cell_34 | cell_35 | cell_36 | cell_37 | cell_38 | cell_39 | cell_40 | cell_41 | cell_42 | cell_43 | cell_44 | cell_45 | cell_46 | cell_47 | cell_48 | cell_49 | cell_50 | cell_51 | cell_52 | cell_53 | cell_54 | cell_55 | cell_56 | cell_57 | cell_58 | cell_59 | cell_60 | cell_61 | cell_62 | cell_63 | cell_64 | cell_65 | cell_66 | cell_67 | cell_68 | cell_69 | cell_70 | cell_71 | cell_72 | cell_73 | cell_74 | cell_75 | cell_76 | cell_77 | cell_78 | cell_79 | cell_80 | cell_81 | cell_82 | cell_83 | cell_84 | cell_85 | cell_86 | cell_87 | cell_88 | cell_89 | cell_90 | cell_91 | cell_92 | cell_93 | cell_94 | cell_95 | cell_96 | cell_97 | cell_98 | cell_99 | cell_100 | cell_101 | cell_102 | cell_103 | cell_104 | cell_105 | cell_106 | cell_107 | cell_108 | cell_109 | cell_110 | cell_111 | cell_112 | cell_113 | cell_114 | cell_115 | cell_116 | cell_117 | cell_118 | cell_119 | cell_120 | cell_121 | cell_122 | cell_123 | cell_124 | cell_125 | cell_126 | cell_127 | cell_128 | cell_129 | cell_130 | cell_131 | cell_132 | cell_133 | cell_134 | cell_135 | cell_136 | cell_137 | cell_138 | cell_139 | cell_140 | cell_141 | cell_142 | cell_143 | cell_144 | cell_145 | cell_146 | cell_147 | cell_148 | cell_149 | cell_150 | cell_151 | cell_152 | cell_153 | cell_154 | cell_155 | cell_156 | cell_157 | cell_158 | cell_159 | cell_160 | cell_161 | cell_162 | cell_163 | cell_164 | cell_165 | cell_166 | cell_167 | cell_168 | cell_169 | cell_170 | cell_171 | cell_172 | cell_173 | cell_174 | cell_175 | cell_176 | cell_177 | cell_178 | cell_179 | cell_180 | cell_181 | cell_182 | cell_183 | cell_184 | cell_185 | cell_186 | cell_187 | cell_188 | cell_189 | cell_190 | cell_191 | cell_192 | cell_193 | cell_194 | cell_195 | cell_196 | cell_197 | cell_198 | cell_199 | cell_200 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gene_1 | 0 | 0 | 66 | 0 | 0 | 70 | 0 | 0 | 0 | 55 | 59 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 58 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 0 | 0 | 0 | 31 | 35 | 0 | 0 | 0 | 57 | 0 | 0 | 0 | 0 | 52 | 0 | 41 | 0 | 0 | 0 | 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 37 | 0 | 0 | 64 | 60 | 0 | 85 | 0 | 0 | 55 | 0 | 0 | 50 | 51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 47 | 0 | 42 | 53 | 43 | 74 | 49 | 69 | 0 | 0 | 44 | 0 | 41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 65 | 0 | 52 | 0 | 0 | 58 | 53 | 0 | 0 | 0 | 49 | 0 | 41 | 0 | 80 | 0 | 0 | 0 | 0 | 48 | 48 | 27 | 0 | 0 | 50 | 63 | 41 | 0 | 0 | 0 | 0 | 60 | 46 | 0 | 55 | 63 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 53 | 0 | 0 | 0 | 71 | 43 | 0 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 54 | 0 | 0 | 45 | 52 | 0 | 0 | 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | 0 | 49 | 0 | 0 | 0 | 0 | 0 |
| gene_2 | 0 | 82 | 0 | 0 | 0 | 0 | 127 | 0 | 89 | 83 | 103 | 0 | 0 | 86 | 84 | 0 | 92 | 77 | 0 | 66 | 110 | 0 | 0 | 0 | 0 | 0 | 65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 97 | 0 | 0 | 0 | 0 | 0 | 0 | 114 | 87 | 0 | 0 | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 83 | 0 | 76 | 0 | 0 | 70 | 0 | 0 | 83 | 0 | 97 | 76 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 123 | 0 | 0 | 0 | 0 | 74 | 0 | 55 | 0 | 0 | 80 | 0 | 0 | 0 | 0 | 0 | 111 | 0 | 0 | 0 | 0 | 77 | 0 | 0 | 0 | 0 | 0 | 81 | 73 | 0 | 0 | 82 | 0 | 0 | 0 | 92 | 0 | 0 | 0 | 71 | 0 | 0 | 0 | 0 | 79 | 0 | 0 | 78 | 0 | 0 | 76 | 0 | 0 | 0 | 75 | 89 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 85 | 0 | 64 | 66 | 86 | 0 | 90 | 0 | 72 | 0 | 0 | 0 | 0 | 0 | 90 | 0 | 100 | 0 | 89 | 0 | 67 | 0 | 77 | 0 | 0 | 0 | 0 | 118 | 77 | 82 | 0 | 0 | 94 | 75 | 0 | 0 | 0 | 82 | 0 | 0 | 0 | 0 | 0 | 69 | 0 | 0 | 0 |
| gene_3 | 0 | 82 | 96 | 0 | 0 | 95 | 0 | 0 | 77 | 77 | 73 | 0 | 0 | 0 | 76 | 0 | 0 | 61 | 0 | 0 | 0 | 0 | 92 | 59 | 0 | 0 | 85 | 0 | 0 | 0 | 50 | 64 | 0 | 0 | 0 | 82 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 84 | 0 | 68 | 80 | 70 | 0 | 66 | 0 | 81 | 0 | 94 | 85 | 0 | 0 | 0 | 68 | 0 | 61 | 94 | 0 | 0 | 0 | 0 | 85 | 0 | 0 | 0 | 62 | 0 | 79 | 71 | 0 | 0 | 0 | 0 | 89 | 0 | 0 | 66 | 0 | 0 | 0 | 0 | 78 | 73 | 0 | 0 | 0 | 75 | 0 | 0 | 0 | 83 | 107 | 0 | 90 | 118 | 0 | 74 | 87 | 79 | 0 | 73 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 86 | 82 | 0 | 0 | 0 | 92 | 0 | 0 | 0 | 0 | 0 | 88 | 0 | 0 | 0 | 0 | 0 | 0 | 75 | 0 | 68 | 0 | 70 | 0 | 0 | 67 | 90 | 76 | 0 | 0 | 89 | 0 | 0 | 0 | 78 | 94 | 0 | 99 | 0 | 0 | 0 | 70 | 0 | 0 | 0 | 84 | 0 | 0 | 0 | 91 | 0 | 109 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 82 | 75 | 97 | 86 | 0 | 89 | 0 | 0 | 0 | 0 | 82 | 87 | 0 | 0 | 0 | 79 | 0 | 0 | 85 | 0 | 62 | 0 | 0 | 0 | 0 | 0 | 0 |
| gene_4 | 0 | 85 | 0 | 88 | 0 | 0 | 0 | 0 | 0 | 0 | 89 | 0 | 0 | 0 | 80 | 0 | 129 | 0 | 0 | 0 | 129 | 0 | 0 | 85 | 0 | 0 | 0 | 47 | 137 | 0 | 155 | 0 | 0 | 0 | 90 | 63 | 0 | 94 | 0 | 0 | 0 | 94 | 46 | 0 | 45 | 0 | 0 | 63 | 0 | 61 | 0 | 0 | 126 | 95 | 90 | 0 | 56 | 68 | 0 | 65 | 0 | 121 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 87 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 25 | 59 | 62 | 0 | 0 | 0 | 0 | 0 | 48 | 42 | 89 | 59 | 66 | 0 | 0 | 0 | 0 | 0 | 0 | 64 | 0 | 83 | 148 | 40 | 97 | 0 | 0 | 0 | 0 | 0 | 91 | 60 | 48 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 77 | 0 | 0 | 0 | 0 | 34 | 0 | 0 | 0 | 0 | 43 | 0 | 0 | 0 | 155 | 0 | 0 | 120 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 111 | 0 | 0 | 0 | 0 | 68 | 61 | 87 | 71 | 75 | 75 | 104 | 0 | 0 | 0 | 144 | 0 | 0 | 0 | 67 | 71 | 104 | 0 | 45 | 0 | 0 | 0 | 0 | 114 | 0 | 57 | 0 | 120 | 91 | 0 | 101 | 69 | 0 | 0 | 0 | 0 | 0 | 0 | 67 | 0 | 0 | 0 | 0 | 70 | 84 | 0 |
| gene_5 | 42 | 0 | 0 | 0 | 69 | 0 | 0 | 0 | 0 | 0 | 0 | 49 | 0 | 0 | 0 | 0 | 63 | 0 | 95 | 0 | 0 | 0 | 0 | 71 | 67 | 0 | 0 | 0 | 50 | 0 | 0 | 0 | 0 | 0 | 56 | 0 | 0 | 69 | 0 | 0 | 0 | 46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 73 | 54 | 0 | 43 | 0 | 0 | 69 | 0 | 0 | 0 | 0 | 0 | 0 | 61 | 0 | 0 | 59 | 0 | 0 | 0 | 0 | 0 | 0 | 45 | 0 | 0 | 64 | 0 | 0 | 69 | 68 | 0 | 54 | 0 | 0 | 66 | 58 | 0 | 55 | 0 | 0 | 0 | 0 | 0 | 0 | 66 | 0 | 0 | 0 | 0 | 0 | 0 | 52 | 0 | 0 | 0 | 0 | 0 | 46 | 0 | 0 | 0 | 0 | 0 | 61 | 56 | 54 | 0 | 43 | 0 | 0 | 0 | 0 | 57 | 63 | 0 | 84 | 74 | 0 | 0 | 46 | 52 | 44 | 0 | 54 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 70 | 0 | 42 | 0 | 0 | 67 | 0 | 0 | 0 | 54 | 65 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 75 | 0 | 0 | 0 | 0 | 58 | 0 | 0 | 0 | 53 | 67 | 0 | 0 | 0 | 0 | 0 | 0 | 71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 64 | 0 | 43 | 0 |
# select valid SNV-gene pairs
# each genotype at least covers 30 cells
# each gene (expression level > 1) at least covers 30 cells
snv.gene.pair.metadata <- eQTLsingle_build_metadata(snvMatrix = toy_snvMatrix,
expressionMatrix = toy_expressionMatrix,
snv.number.of.cells = 30,
expression.min = 1,
expression.number.of.cells = 30)Format of snv.gene.pair.metadata,
SNVid: Id of SNV, represented as CHR__POS;
Num_cells_ref: Num_cells_ref: number of cells in REF group;
Num_cells_alt: Num_cells_alt: number of cells in ALT group;
Ref_cells: cell list of REF group;
Alt_cells: cell list of ALT group;
GeneList: gene list, these genes will be tested with the SNV in this row;
Num_gene: number of genes in gene list;
CellList: whole cell list of REF and ALT group.
实例操作
Step 2: Conduct eQTL analysis on valid SNV-gene pairs.
# conduct eQTL analysis on valid SNV-gene pairs
eQTL.result <- eQTLsingle(toy_expressionMatrix,
snv.gene.pair.metadata)# Suppose we are interested at the pair between
# SNV with snvid "SNV_3"
# gene with geneid "gene_16"Format of eQTL.result,
SNVid: Id of SNV, represented as CHR__POS;
Geneid: gene id of target gene;
sample_size_Ref, sample_size_Alt: number of cells in REF and ALT group;
theta_Ref, theta_Alt, mu_Ref, mu_Alt, size_Ref, size_Alt, prob_Ref, prob_Alt: estimated parameters of ZINB for REF group and ALT group;
total_mean_Ref, total_mean_Alt: mean of the REF group and ALT group;
foldChange: fold change of mean of REF group (total_mean_Ref) with respect to mean of ALT group (total_mean_Alt) chi2LR1: chi square statistic for the test;
pvalue, adjusted_pvalue: pvalue and adjusted pvalue of the test. If adjusted p-value is smaller than some threshold, this SNV shows significant eQTL effect on the target gene;
Remark: to record abnormity
Step 3: Visualize eQTL effect
# Suppose we are interested at the pair between
# SNV with snvid "SNV_3"
# gene with geneid "gene_16"
# select out a SNV-gene pair
eQTL.result.target <- eQTL.result[(eQTL.result$SNVid == "SNV_3") &
(eQTL.result$Geneid == "gene_16"),]
kable(eQTL.result.target[1:5,])| SNVid | Geneid | sample_size_Ref | sample_size_Alt | theta_Ref | theta_Alt | mu_Ref | mu_Alt | size_Ref | size_Alt | prob_Ref | prob_Alt | total_mean_Ref | total_mean_Alt | foldChange | chi2LR1 | pvalue | adjusted_pvalue | Remark | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | SNV_3 | gene_16 | 91 | 89 | 0.032967 | 0.0449438 | 59.94318 | 291.1177 | 9.247933 | 12.17195 | 0.1336578 | 0.0401331 | 57.96703 | 278.0337 | 0.2084892 | 337.0705 | 0 | 0 | NA |
| NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA.1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA.2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| NA.3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
# to draw violin plots of gene expression in both REF group and ALT group
eQTLsingle_visualization(eQTL.result.target,toy_expressionMatrix, snv.gene.pair.metadata)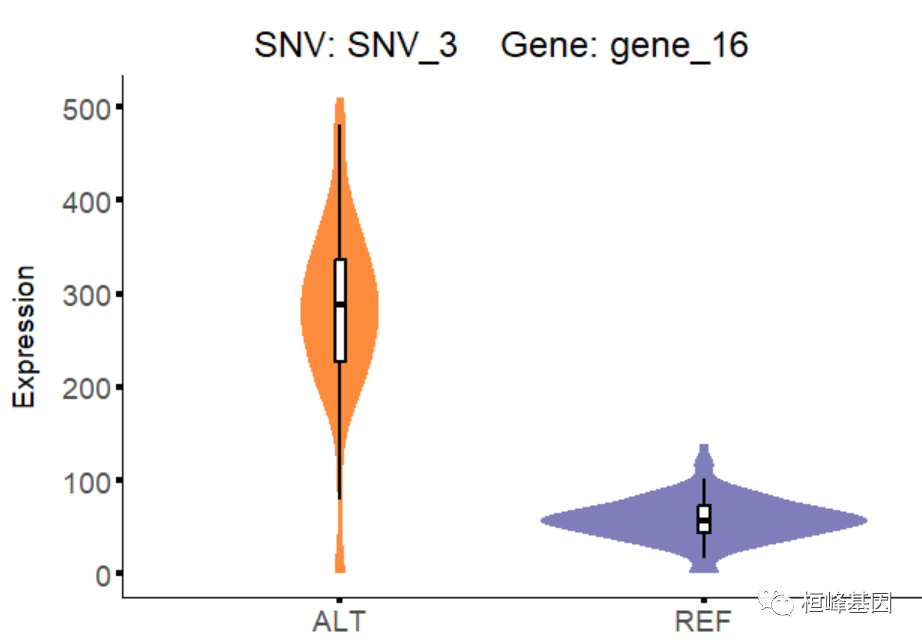
Raw-data mode
这个可以参考网站,这时就需要有一定 shell 基础的同学,并且对一些生物学软件很熟练,但是还算好做,思路很清晰。
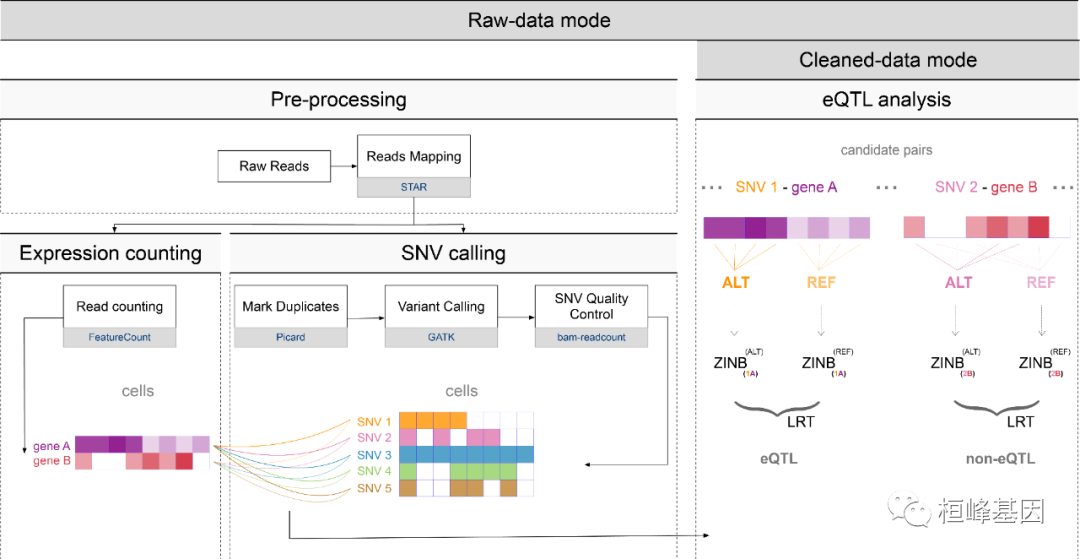
涉及到的软件:
samtoolsfeatureCountsGATKpicardjavabam-readcountPython3Pandas
如果您只有FASTQ数据,并且希望使用我们的脚本生成表达式矩阵和SNV矩阵,则可以运行存储在。/inst/中的shell脚本。可以直接从R环境中执行这些脚本,但我们建议您在shell中使用,主要为两部,后面的就可以直接读入软件包 eQTLsingle 中操作了。
预处理:从FASTQ数据中生成突变谱和表达谱;
SNV质量控制:对选中的SNV进行读取计数,确认有效的突变,以便进行进一步的eQTL分析。
后续分析我们可以结合一些临床预测的方法,比如文章中的后续分析 Discoveries on the rs3169950-PSMB6 cis-eQTL pair in gliomaspheres。
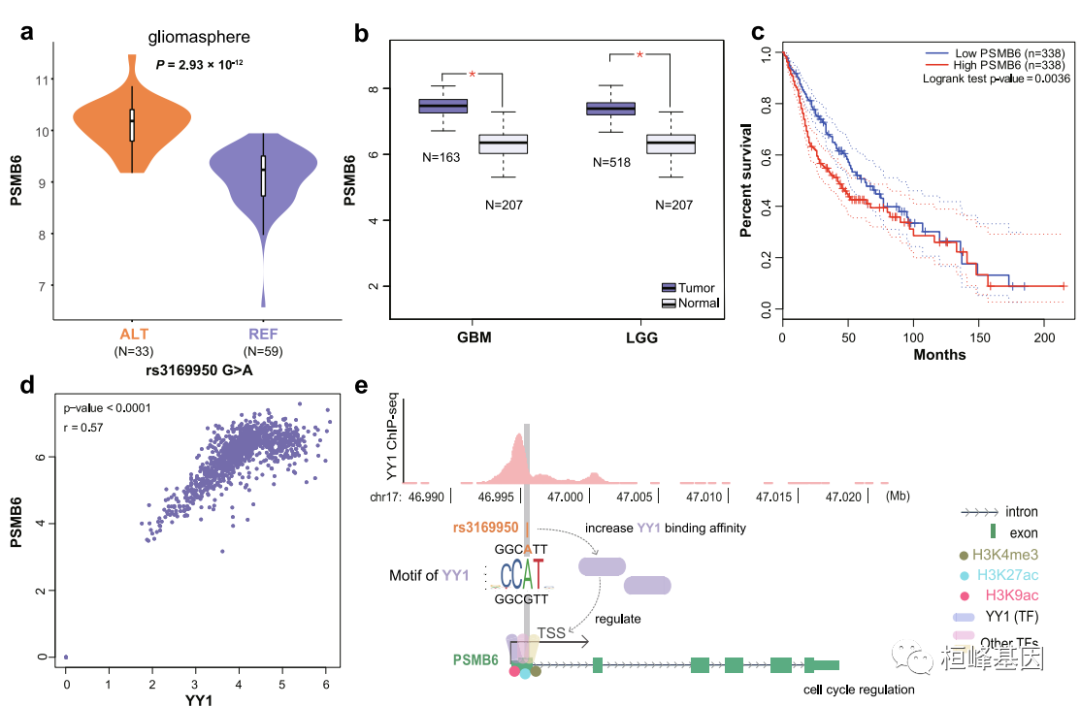
(a)胶质球中REF/ALT型细胞rs3169950位点PSMB6的表达水平 (bonferroni校正p值= 2.93 × 10−12)。
(b) PSMB6基因表达与 基于TCGA和GTEx数据的脑肿瘤(GBM和LGG)和正常脑样本。红色星号表示单因素方差分析检验p值≤0.05。
(c)按PSMB6基因表达水平分层的TCGA GBM和LGG患者Kaplan-Meier生存图患者分为低表达组和 高表达组以PSMB6表达水平中位数作为截止值。
(d) GTEx脑数据中YY1与PSMB6基因表达的相关性。Pearson相关系数为0.57 (p值< 0.001)。
(e) PSMB6上转录因子YY1的ChIP-seq信号和rs3169950调控PSMB6的假设模型 表达式。
Reference
Ma T, Li H, Zhang X. Discovering single-cell eQTLs from scRNA-seq data only. Gene. 2022;829:146520. doi:10.1016/j.gene.2022.146520
利用 Cleaned-data mode 还是很快分析出来,但是从 Raw-data mode 开始做还是要求有一定生信分析基础的,有需求的老师可以联系桓峰基因,关注桓峰基因公众号,轻松学生信,高效发文章!
桓峰基因,铸造成功的您!
未来桓峰基因公众号将不间断的推出单细胞系列生信分析教程,
敬请期待!!
桓峰基因官网正式上线,请大家多多关注,还有很多不足之处,大家多多指正!
http://www.kyohogene.com/
桓峰基因和投必得合作,文章润色优惠85折,需要文章润色的老师可以直接到网站输入领取桓峰基因专属优惠券码:KYOHOGENE,然后上传,付款时选择桓峰基因优惠券即可享受85折优惠哦!https://www.topeditsci.com/























 5100
5100











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








