Jun Ma, Yuting He, Feifei Li, Lin Han, Chenyu You, and Bo Wang
Abstract
摘要
In this study, we present MedSAM, the inaugural foundation model designed for universal medical image segmentation. Harnessing the power of a meticulously curated dataset comprising over one million images, MedSAM not only outperforms existing state-of-the-art segmentation foundation models, but also exhibits comparable or even superior performance to specialist models.
在本研究中,我们提出了MedSAM,设计为通用医学图像分割的第一个基础模型。MedSAM利用一个精心策划的包含超过100万张图像的数据集的力量,不仅优于现有的最先进的分割基础模型,而且表现出与专家模型相当甚至更优越的性能。
I
NTRODUCTIO
N
介绍
Motivated by the remarkable generality of the Segment Anything Model (SAM) [4], we introduce MedSAM, the first foundation model for universal medical image segmentation. MedSAM is adapted from the SAM model on an unprecedented scale, with more than one million medical image-mask pairs. We thoroughly evaluate MedSAM through comprehensive experiments on over 70 internal validation tasks and 40 external validation tasks, spanning a variety of anatomical structures, pathological conditions, and medical imaging modalities. Experimental results demonstrate that MedSAM consistently outperforms the state-of-the-art (SOTA) segmentation foundation model, while achieving performance on par with, or even surpassing specialist models. These results highlight the potential of MedSAM as a powerful tool for medical image segmentation.
基于分割万物模型(SAM)[4]的显著通用性,我们引入了MedSAM,即通用医学图像分割的第一个基础模型。MedSAM改编SAM模型的规模是前所未有的,有超过100万对医学图像mask。我们通过对70多个内部验证任务和40个外部验证任务的综合实验,彻底评估了MedSAM,包括各种解剖结构、病理条件和医学成像模式。实验结果表明,MedSAM的性能始终优于最先进的(SOTA)分割基础模型,同时达到与专业模型相同,甚至超过的性能。这些结果突出了MedSAM作为医学图像分割的强大工具的潜力。
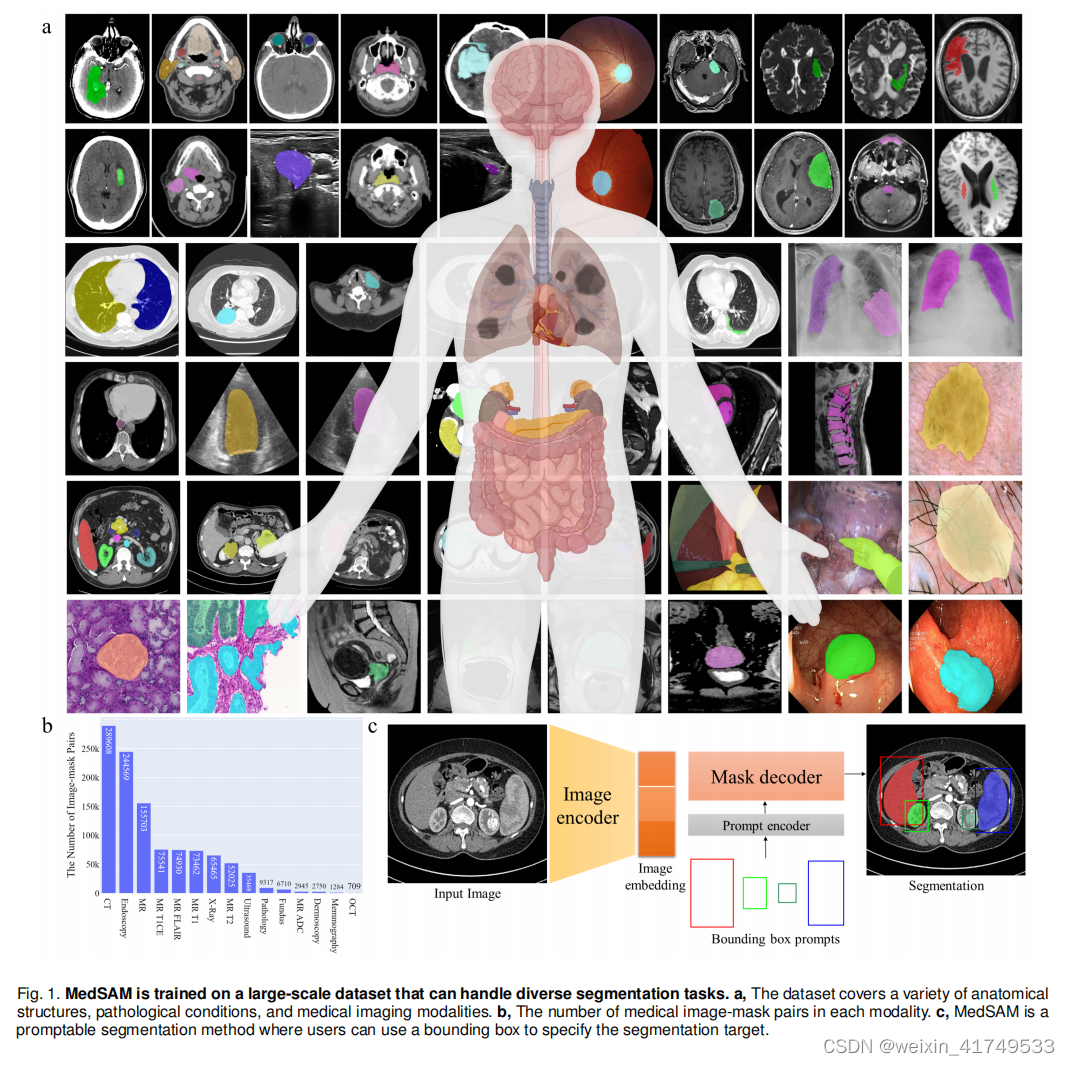
We introduce MedSAM, a deep learning-powered foundation model designed for the segmentation of a wide array of anatomical structures and lesions across diverse medical imaging modalities. MedSAM is trained on a meticulously assembled large-scale dataset comprising over one million medical image-mask pairs. Its promptable configuration strikes
an optimal balance between automation and customization, rendering MedSAM a versatile tool for universal medical image segmentation.
我们介绍了MedSAM,这是一种深度学习驱动的基础模型,设计用于分割不同医学成像模式中广泛的解剖结构和病变。MedSAM是在一个精心组装的由超过100万对医学image-mask组成的大规模数据集上进行训练的。它的快速配置在自动化和定制之间达到了最佳的平衡,使MedSAM成为通用医疗图像分割的通用工具。
Dataset curation and pre-processing
数据集管理以及预处理
We curated a comprehensive dataset by collating images from publicly available medical image segmentation datasets, which were obtained from various sources across the internet. These sources include The Cancer Imaging Archive (TCIA) at https://www.cancerimagingarchive.net/, Kaggle at https://www.kaggle.com/, Grand-Challenge at https:// grand-challenge.org/challenges/, Scientific Data at https://www.nature.com/sdata/, CodaLab at https://codalab.lisn. upsaclay.fr/, and segmentation challenges within the Medical Image Computing and Computer Assisted Intervention Society (MICCAI) at http://www.miccai.org/. The complete list of datasets utilized is presented in Supplementary Table 1-4
我们通过整理来自公开的医学图像分割数据集的图像来整理一个全面的数据集,这些数据集从互联网上的各种来源获得。这些资料来源包括https://www.cancerimagingarchive.net/的癌症成像档案(TCIA),https://www.kaggle.com/的Kaggle,https://grand-challenge.org/challenges/的大挑战,https://www.nature.com/sdata/的科学数据,https://codalab.lisn的CodaLab。upsaclay.fr/,以及在http://www.miccai.org/的医学图像计算和计算机辅助干预协会(MICCAI)内的分割挑战。所使用的数据集的完整列表见补充表1-4
数据预处理部分主要是分别对三维、二维的数据都做了归一化以及统一size等的操作,详见论文细节
Network architecture
网络结构
To strike a balance between segmentation performance and computational efficiency, we employed the base ViT model as the image encoder since extensive evaluation indicated that larger ViT models, such as ViT Large and ViT Huge, offered only marginal accuracy improvements [4] while significantly increasing computational demands.
为了在分割性能和计算效率之间取得平衡,我们使用了基础ViT模型作为图像编码器,因为广泛的评估表明,更大的ViT模型,如ViT Large和ViT Huge,只提供了7个边际精度改进[4],同时显著增加了计算需求.
. Specifically, the base ViT model consists of 12 transformer layers [9], with each block comprising a multi-head self-attention block and a Multilayer Perceptron (MLP) block incorporating layer normalization [17]. Pre-training was performed using masked auto-encoder modeling [18], followed by fully supervised training on the SAM dataset [4]. The input image (
1024
×
1024
×
3) was reshaped into a sequence of flattened 2D patches with the size
16
×
16
×
3, yielding a feature size in image embedding of 64
×64 after passing through the image encoder, which is 16× downscaled. The prompt encoders mapped the corner point of the bounding box prompt to 256-dimensional vectorial embeddings [8]. In particular, each bounding box was represented by an embedding pair of the top-left corner point and the bottom-right corner point. To facilitate real-time user interactions once the image embedding had been computed, a lightweight mask decoder architecture was employed. It comprised two transformer layers [9] for fusing the image embedding and prompt encoding, and two transposed convolutional layers to enhance the embedding resolution to
256
×
256. Subsequently, the embedding underwent sigmoid activation, followed by bi-linear interpolations to match the input size.
具体来说,基础ViT模型由12个变压器层[9]组成,每个块包括一个多头自注意块和包含层归一化[17]的多层感知器(MLP)块。使用掩码自动编码器建模[18]进行预训练,然后对SAM数据集[4]进行完全监督训练。将输入图像(1024×1024×3)重塑为大小为16×16×3的扁平2维补丁序列,经过图像编码器后,得到图像嵌入64×64的特征尺寸,缩小16×。提示符编码器将边界框提示符的角点映射到256维向量嵌入[8]。特别是,每个边界框都由左上角点和右下角点的嵌入对表示。为了便于计算图像嵌入后的实时用户交互,采用了轻量级掩码解码器架构。它包括两个变压器层[9],以融合图像嵌入和提示编码,以及两个转置的卷积层,以提高嵌入分辨率到256×256。随后,嵌入进行s型激活,然后进行双线性插值以匹配输入大小。
Training
训练
The model was initialized with the pre-trained SAM model with the ViT-Base model. We fix the prompt encoder since it can already encode the bounding box prompt. All the trainable parameters in the image encoder and mask decoder were updated during training. Specifically, the number of trainable parameters for the image encoder and mask decoder are 89,670,912 and 4,058,340, respectively. The bounding box prompt was simulated from the ground-truth mask with a random perturbation of 0-20 pixels. The loss function is the unweighted sum between Dice loss and cross-entropy loss, which has been proven to be robust in various segmentation tasks [3]. Specifically, let
S, G denote the segmentation result and ground truth, respectively.
s
i
, g
i
denote the predicted segmentation and ground truth of voxel
i
, respectively.
N is the number of voxels in the image
I
. Cross-entropy loss is defined by

该模型采用预先训练好的SAM模型和ViT-Base模型进行初始化。我们修复了提示符编码器,因为它已经可以编码边界框提示符。在训练过程中,对图像编码器和掩码解码器中的所有可训练参数都进行了更新。具体地说,图像编码器和掩码解码器的可训练参数数分别为89、670、912和4,058、340。边界框提示是由0-20像素的地面真实掩模的随机扰动模拟的。损失函数是DICE损失和交叉熵损失之间的非加权和,在各种分割任务[3]中被证明是鲁棒性。具体来说,设S、G分别表示分割结果和groundtruth。si,gi分别表示体素i的预测分割和地面真值。N为图像i中的体素数,交叉熵损失定义如图。
Evaluation metrics
评估矩阵
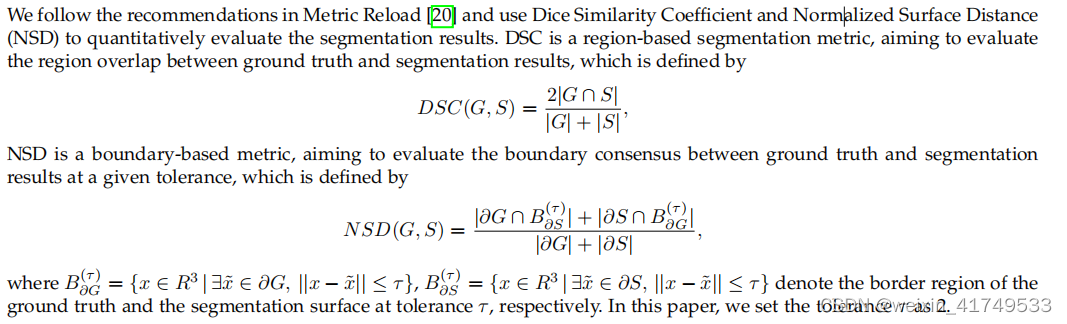
DSC,NSD可以自行百度。
Code availability
开源代码
All code was implemented in Python (3.10) using Pytorch (2.0) as the base deep learning framework. We also used several python packages for data analysis and results visualization, including SimpleITK (2.2.1), nibabel (5.1.0), torchvision (0.15.2), numpy (1.24.3), scikit-image (0.20.0), opencvpython (4.7.0), scipy (1.10.1), and pandas (2.0.2), matplotlib (3.7.1), and plotly
(5.15.0). Biorender was used to create Fig. 1a. The training script, inference script, and trained model have been publicly available at https://github.com/bowang-lab/MedSAM.





















 1759
1759











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








