最近想学一学宏基因组的分箱工具使用(讲真的,感觉bin还是挺复杂的,不是我这种小白该去涉猎的),本来想看看老牌工具metaWRAP的使用细节,奈何微信推送了一条新的分箱工具——SemiBin,还是基于当下很热门的半监督+神经网络的模型构建的,感觉很新鲜,想尝尝鲜,于是就去SemiBin的github上学习一下基本用法:
该软件跟着install的引导去下载就可以了,还是比较简单,不过其中有几个较大的配置文件/软件要下载,望大家耐心等待。另外需要注意的是组装工具megahit,比对工具bowtie2以及samtools需要自己在SemiBin所在的环境中下载,具体的下载安装细节搬运一下github:
# semibin的下载和安装
conda create -n SemiBin
conda activate SemiBin
conda install -c conda-forge -c bioconda semibin
# 辅助semibin的软件下载
conda install -c bioconda bowtie2
conda install -c bioconda samtools
conda install -c bioconda megahit
# 检查semibin是否安装好安装
SemiBin check_install
# 出现下面这样就没什么问题了,表明安装成功:
Looking for dependencies...
bedtools : /home/dell/miniconda3/envs/SemiBin/bin/bedtools
hmmsearch : /home/dell/miniconda3/envs/SemiBin/bin/hmmsearch
mmseqs : /home/dell/miniconda3/envs/SemiBin/bin/mmseqs
FragGeneScan : /home/dell/miniconda3/envs/SemiBin/bin/FragGeneScan
prodigal : /home/dell/miniconda3/envs/SemiBin/bin/prodigal
Installation OK
If you find SemiBin useful, please cite:
Pan, S., Zhu, C., Zhao, XM. et al. A deep siamese neural network improves metagenome-assembled genomes in microbiome datasets across different environments. Nat Commun 13, 2326 (2022). https://doi.org/10.1038/s41467-022-29843-y.
下载好后,就是实战了,从说明文档中看该软件的使用较为简单,应该是基于最理想的情况,我就以最简单的单样本、简单bin为例进行尝试:
实例文件可以在github上找:GitHub - BigDataBiology/SemiBin_tutorial_from_scratch
点到single_sample_binning这个文件夹中,可见里面有一个样本的双端测序的文件

ok,在linux端的操作是:
# 1. 下载实例数据集,我是采用手动下载并上传的方式
# 2. cd到 single_sample_binning文件夹下
cd single_sample_binning
# 3. 组装
megahit -1 sample1_R1.fastq.gz \
-2 sample1_R2.fastq.gz \
--out-dir assembly_contig \
--out-prefix R1
# 4. mapping 并进行binning前处理:
bowtie2-build \
-f assembly_contig/R1.contigs.fa assembly_contig/R1.contigs.fa
bowtie2 -q --fr \
-x assembly_contig/R1.contigs.fa \
-1 sample1_R1.fastq.gz \
-2 sample1_R2.fastq.gz \
-S sample1.sam \
-p 64
samtools view -h -b -S sample1.sam -o sample1.bam -@ 64
samtools view -b -F 4 sample1.bam -o sample1.mapped.bam -@ 64
samtools sort \
-m 1000000000 sample1.mapped.bam \
-o sample1.mapped.sorted.bam -@ 64
# 5. Easy single binning mode 最简单的binning方式
SemiBin single_easy_bin \
-i assembly_contig/R1.contigs.fa \
-b sample1.mapped.sorted.bam \
-o easy_single_sample_output
# ========================================================
# 在这边出问题了:
(SemiBin) dell 20:18:05 /home/DB/fqz/mydata/SemiBin_testdata/single_sample_binning
$ SemiBin single_easy_bin \
> -i assembly_contig/R1.contigs.fa \
> -b sample1.mapped.sorted.bam \
> -o easy_single_sample_output
2022-08-25 20:19:36,299 - Setting number of CPUs to 112
2022-08-25 20:19:36,300 - Do not detect GPU. Running with CPU.
2022-08-25 20:19:36,315 - Generate training data.
2022-08-25 20:19:37,052 - Calculating coverage for every sample.
2022-08-25 20:19:37,057 - Processing `sample1.mapped.sorted.bam`
2022-08-25 20:19:37,805 - Processed:sample1.mapped.sorted.bam
2022-08-25 20:19:37,873 - Start generating kmer features from fasta file.
2022-08-25 20:19:39,087 - Running mmseqs and generate cannot-link file.
2022-08-25 20:19:39,149 - Downloading GTDB to /home/dell/.cache/SemiBin/mmseqs2-GTDB. It will take a while..
# 第一使用该工具会下载软件依赖的GTDB数据库,由于放在外网所以下载速度很感人.....多次下载失败
直接使用SemiBin自带的数据库下载函数也不行,不过给了提示,该软件依赖的GTDB数据库使用的版本是v95:

所以,本次尝试主要遇到的问题就是SemiBin依赖的GTDB数据库下载总是失败,应该是外网限速的问题,我不会FQ,也没钱整DL,所以只能自己想办法,于是再次采用手动下载手动上传并安装GTDB v95版本的方式,首先去GTDB数据库的网站找v95版本,然后下载上传到 /home/dell/.cache/SemiBin/mmseqs2-GTDB 这个文件夹下,再次运行 SemiBin single_easy_bin 命令,结果文件被直接覆盖(即重新龟速下载),后来将手动下载的文件名命名成软件需要的名字,还是被覆盖,重复前述步骤,并将手动下载的GTDB数据库解压缩,还是被覆盖。所以得出的结论是这个文件可能不是SemiBin能够识别的文件,版本对,但是不能被识别,说明不是这个文件,同样还是中断龟速下载后,报错文件给了我提示:
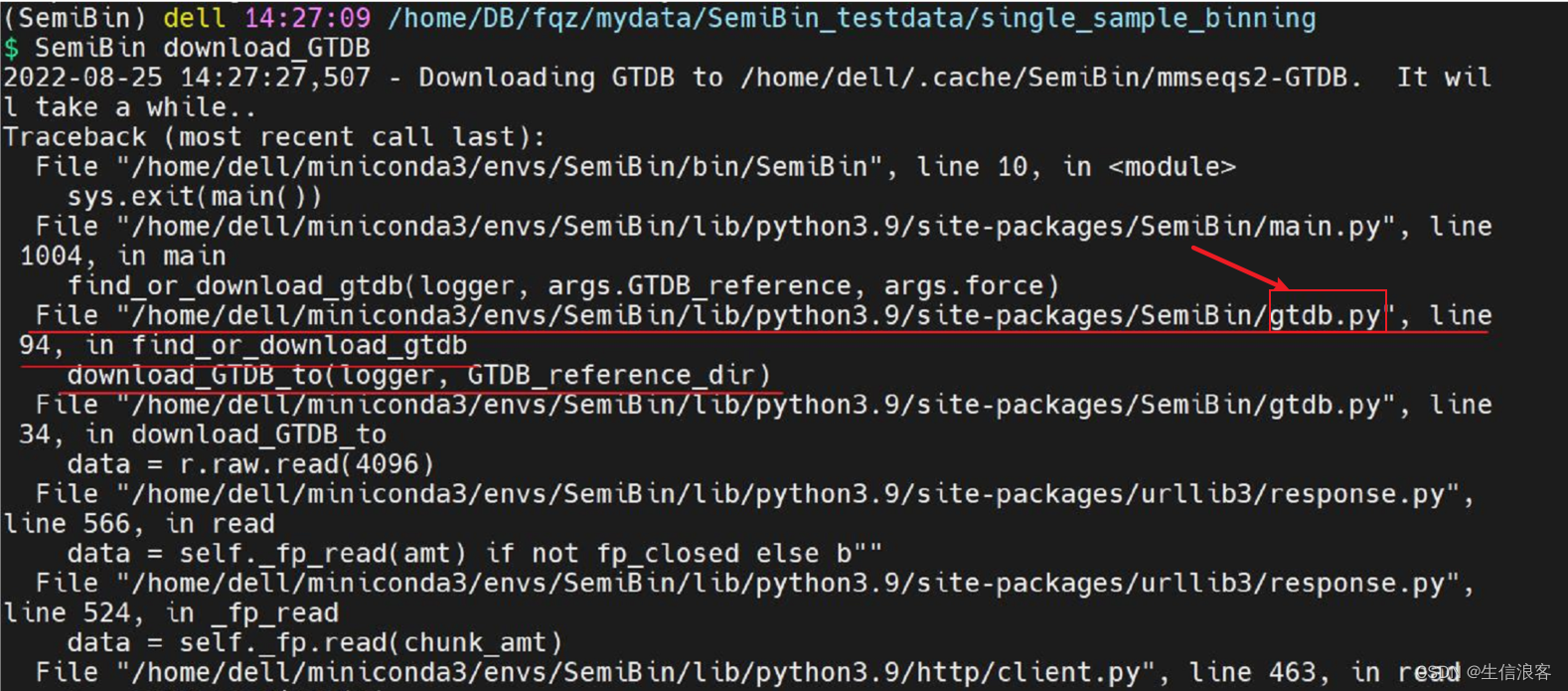
gtdb.py这个python脚本应该与下载GTBD数据库有关,于是去找github主页该文件的位置:

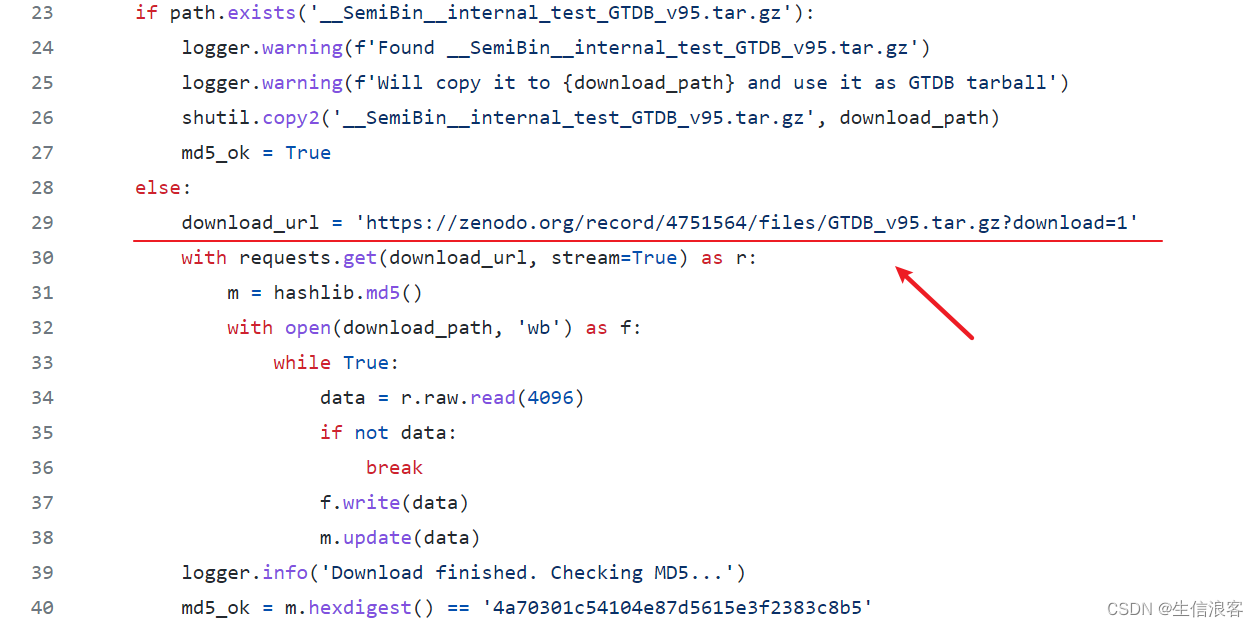
还好作者在zenodo平台上留了一个GTDB数据库v95版本的备份,不然还真难搞
GTDB reference genome generated by MMseqs2 used in SemiBin. | Zenodo
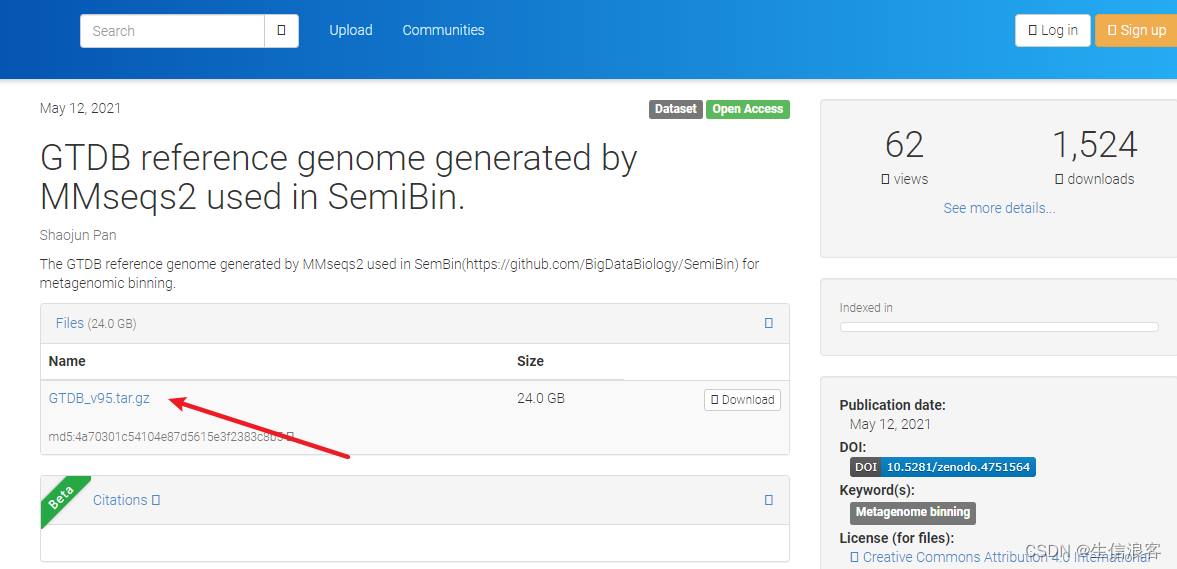
下载链接: https://zenodo.org/record/4751564/files/GTDB_v95.tar.gz?download=1
基于这个下载链接,使用迅雷,在保存到迅雷云盘之后本地下载的方式,我感觉比较快
下载完毕后,上传到指定位置(我的位置是~/.cache/SemiBin/mmseqs2-GTDB),然后解压缩!一定要解压缩,不然SemiBin还不识别然后将你的文件覆盖掉重新下载......
解压缩就随意了,我是使用pigz + tar的方式解压:
pigz -d GTDB_v95.tar.gz
tar -xf GTDB_v95.tar
# 也可以直接 tar -zxvf GTDB_v95.tar.gz
# 验证一下是否已经下载好,semibin能够识别:
SemiBin download_GTDB
2022-08-25 20:36:01,669 - Found GTDB directory: `/home/dell/.cache/SemiBin/mmseqs2-GTDB`.
If you find SemiBin useful, please cite:
Pan, S., Zhu, C., Zhao, XM. et al. A deep siamese neural network improves metagenome-assembled genomes in microbiome datasets across different environments. Nat Commun 13, 2326 (2022). https://doi.org/10.1038/s41467-022-29843-y.
# 可以继续了查看解压后文件:
再次运行单样本简单binning,没有报错,一切正常了:
(SemiBin) dell 20:36:42 /home/DB/fqz/mydata/SemiBin_testdata/single_sample_binning
$ SemiBin single_easy_bin \
> -i assembly_contig/R1.contigs.fa \
> -b sample1.mapped.sorted.bam \
> -o easy_single_sample_output
# 下面全是软件binning过程输出:
2022-08-25 20:37:06,248 - Setting number of CPUs to 112
2022-08-25 20:37:06,249 - Do not detect GPU. Running with CPU.
2022-08-25 20:37:06,264 - Generate training data.
2022-08-25 20:37:06,920 - Calculating coverage for every sample.
2022-08-25 20:37:06,925 - Processing `sample1.mapped.sorted.bam`
2022-08-25 20:37:07,667 - Processed:sample1.mapped.sorted.bam
2022-08-25 20:37:07,721 - Start generating kmer features from fasta file.
2022-08-25 20:37:08,903 - Running mmseqs and generate cannot-link file.
2022-08-25 20:37:08,915 - Found GTDB directory: `/home/dell/.cache/SemiBin/mmseqs2-GTDB`.
createdb /tmp/tmp5y44n4h6/SemiBinFiltered.fa /tmp/tmp5y44n4h6/contig_DB
MMseqs Version: 13.45111
Database type 0
Shuffle input database true
Createdb mode 0
Write lookup file 1
Offset of numeric ids 0
Compressed 0
Verbosity 3
Converting sequences
[304] 0s 9ms
Time for merging to contig_DB_h: 0h 0m 0s 19ms
Time for merging to contig_DB: 0h 0m 0s 6ms
Database type: Nucleotide
Time for processing: 0h 0m 0s 45ms
taxonomy /tmp/tmp5y44n4h6/contig_DB /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB easy_single_sample_output/mmseqs_contig_annotation/mmseqs_contig_annotation /tmp/tmp5y44n4h6 --tax-lineage 1 --threads 112
MMseqs Version: 13.45111
ORF filter 1
ORF filter e-value 100
ORF filter sensitivity 2
LCA mode 3
Taxonomy output mode 0
Majority threshold 0.5
Vote mode 1
LCA ranks
Column with taxonomic lineage 1
Compressed 0
Threads 112
Verbosity 3
Taxon blacklist 12908:unclassified sequences,28384:other sequences
Substitution matrix nucl:nucleotide.out,aa:blosum62.out
Add backtrace false
Alignment mode 1
Alignment mode 0
Allow wrapped scoring false
E-value threshold 1
Seq. id. threshold 0
Min alignment length 0
Seq. id. mode 0
Alternative alignments 0
Coverage threshold 0
Coverage mode 0
Max sequence length 65535
Compositional bias 1
Max reject 5
Max accept 30
Include identical seq. id. false
Preload mode 0
Pseudo count a 1
Pseudo count b 1.5
Score bias 0
Realign hits false
Realign score bias -0.2
Realign max seqs 2147483647
Gap open cost nucl:5,aa:11
Gap extension cost nucl:2,aa:1
Zdrop 40
Seed substitution matrix nucl:nucleotide.out,aa:VTML80.out
Sensitivity 2
k-mer length 0
k-score 2147483647
Alphabet size nucl:5,aa:21
Max results per query 300
Split database 0
Split mode 2
Split memory limit 0
Diagonal scoring true
Exact k-mer matching 0
Mask residues 1
Mask lower case residues 0
Minimum diagonal score 15
Spaced k-mers 1
Spaced k-mer pattern
Local temporary path
Rescore mode 0
Remove hits by seq. id. and coverage false
Sort results 0
Mask profile 1
Profile E-value threshold 0.001
Global sequence weighting false
Allow deletions false
Filter MSA 1
Maximum seq. id. threshold 0.9
Minimum seq. id. 0
Minimum score per column -20
Minimum coverage 0
Select N most diverse seqs 1000
Min codons in orf 30
Max codons in length 32734
Max orf gaps 2147483647
Contig start mode 2
Contig end mode 2
Orf start mode 1
Forward frames 1,2,3
Reverse frames 1,2,3
Translation table 1
Translate orf 0
Use all table starts false
Offset of numeric ids 0
Create lookup 0
Add orf stop false
Overlap between sequences 0
Sequence split mode 1
Header split mode 0
Chain overlapping alignments 0
Merge query 1
Search type 0
Search iterations 1
Start sensitivity 4
Search steps 1
Exhaustive search mode false
Filter results during exhaustive search 0
Strand selection 1
LCA search mode false
Disk space limit 0
MPI runner
Force restart with latest tmp false
Remove temporary files false
extractorfs /tmp/tmp5y44n4h6/contig_DB /tmp/tmp5y44n4h6/10505879959436434455/orfs_aa --min-length 30 --max-length 32734 --max-gaps 2147483647 --contig-start-mode 2 --contig-end-mode 2 --orf-start-mode 1 --forward-frames 1,2,3 --reverse-frames 1,2,3 --translation-table 1 --translate 1 --use-all-table-starts 0 --id-offset 0 --create-lookup 0 --threads 112 --compressed 0 -v 3
[=================================================================] 100.00% 392 0s 61ms
Time for merging to orfs_aa_h: 0h 0m 0s 48ms
Time for merging to orfs_aa: 0h 0m 0s 60ms
Time for processing: 0h 0m 0s 299ms
prefilter /tmp/tmp5y44n4h6/10505879959436434455/orfs_aa /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/orfs_pref --sub-mat nucl:nucleotide.out,aa:blosum62.out --seed-sub-mat nucl:nucleotide.out,aa:VTML80.out -s 2 -k 0 --k-score 2147483647 --alph-size nucl:5,aa:21 --max-seq-len 65535 --max-seqs 1 --split 0 --split-mode 2 --split-memory-limit 0 -c 0 --cov-mode 0 --comp-bias-corr 1 --diag-score 0 --exact-kmer-matching 0 --mask 1 --mask-lower-case 0 --min-ungapped-score 3 --add-self-matches 0 --spaced-kmer-mode 1 --db-load-mode 0 --pca 1 --pcb 1.5 --threads 112 --compressed 0 -v 3
Query database size: 31958 type: Aminoacid
Estimated memory consumption: 753G
Target database size: 106052079 type: Aminoacid
Index table k-mer threshold: 163 at k-mer size 7
Index table: counting k-mers
[=================================================================] 100.00% 106.05M 2m 3s 49ms
Index table: Masked residues: 269144627
Index table: fill
[=================================================================] 100.00% 106.05M 2m 31s 544ms
Index statistics
Entries: 28079084193
DB size: 170435 MB
Avg k-mer size: 21.936785
Top 10 k-mers
SGQQRIA 397575
GPGGKLL 319073
GGQRVAR 221105
YTGTGKG 177317
LSGQQAI 153681
GGRRVAR 125622
ALGNGKS 109876
LLGPGKT 107267
GRFVVEV 105507
TPHDFEV 88676
Time for index table init: 0h 4m 54s 119ms
Process prefiltering step 1 of 1
k-mer similarity threshold: 163
Starting prefiltering scores calculation (step 1 of 1)
Query db start 1 to 31958
Target db start 1 to 106052079
[=================================================================] 100.00% 31.96K 0s 537ms
30.136279 k-mers per position
27916 DB matches per sequence
0 overflows
0 queries produce too many hits (truncated result)
0 sequences passed prefiltering per query sequence
0 median result list length
29412 sequences with 0 size result lists
Time for merging to orfs_pref: 0h 0m 0s 21ms
Time for processing: 0h 5m 16s 25ms
rescorediagonal /tmp/tmp5y44n4h6/10505879959436434455/orfs_aa /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/orfs_pref /tmp/tmp5y44n4h6/10505879959436434455/orfs_aln --sub-mat nucl:nucleotide.out,aa:blosum62.out --rescore-mode 2 --wrapped-scoring 0 --filter-hits 0 -e 100 -c 0 -a 0 --cov-mode 0 --min-seq-id 0 --min-aln-len 0 --seq-id-mode 0 --add-self-matches 0 --sort-results 0 --db-load-mode 0 --threads 112 --compressed 0 -v 3
[=================================================================] 100.00% 31.96K 0s 63ms
Time for merging to orfs_aln: 0h 0m 0s 10ms=================> ] 91.58% 29.27K eta 0s
Time for processing: 0h 0m 5s 87ms
createsubdb /tmp/tmp5y44n4h6/10505879959436434455/orfs_aln.list /tmp/tmp5y44n4h6/10505879959436434455/orfs_aa /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter --subdb-mode 1 -v 3
Time for merging to orfs_filter: 0h 0m 0s 0ms
Time for processing: 0h 0m 0s 22ms
rmdb /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter_h -v 3
Time for processing: 0h 0m 0s 2ms
createsubdb /tmp/tmp5y44n4h6/10505879959436434455/orfs_aln.list /tmp/tmp5y44n4h6/10505879959436434455/orfs_aa_h /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter_h --subdb-mode 1 -v 3
Time for merging to orfs_filter_h: 0h 0m 0s 0ms
Time for processing: 0h 0m 0s 22ms
Create directory /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy
taxonomy /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/orfs_tax /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy --tax-output-mode 2 --tax-lineage 0 --threads 112 --alignment-mode 1 -e 1 --max-rejected 5 --max-accept 30 -s 2 --spaced-kmer-mode 1 --min-length 30 --max-length 32734 --orf-start-mode 1
Create directory /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/tmp_hsp1
search /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/first /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/tmp_hsp1 --alignment-mode 1 -e 1 --max-rejected 5 --max-accept 30 --threads 112 -s 2 --spaced-kmer-mode 1 --min-length 30 --max-length 32734 --orf-start-mode 1 --lca-search 1
prefilter /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/tmp_hsp1/10382565990596377146/pref_0 --sub-mat nucl:nucleotide.out,aa:blosum62.out --seed-sub-mat nucl:nucleotide.out,aa:VTML80.out -k 0 --k-score 2147483647 --alph-size nucl:5,aa:21 --max-seq-len 65535 --max-seqs 300 --split 0 --split-mode 2 --split-memory-limit 0 -c 0 --cov-mode 0 --comp-bias-corr 1 --diag-score 1 --exact-kmer-matching 0 --mask 1 --mask-lower-case 0 --min-ungapped-score 15 --add-self-matches 0 --spaced-kmer-mode 1 --db-load-mode 0 --pca 1 --pcb 1.5 --threads 112 --compressed 0 -v 3 -s 2.0
Query database size: 2531 type: Aminoacid
Estimated memory consumption: 753G
Target database size: 106052079 type: Aminoacid
Index table k-mer threshold: 163 at k-mer size 7
Index table: counting k-mers
[=================================================================] 100.00% 106.05M 2m 4s 599ms
Index table: Masked residues: 269144627
Index table: fill
[=================================================================] 100.00% 106.05M 2m 32s 452ms
Index statistics
Entries: 28079084193
DB size: 170435 MB
Avg k-mer size: 21.936785
Top 10 k-mers
SGQQRIA 397575
GPGGKLL 319073
GGQRVAR 221105
YTGTGKG 177317
LSGQQAI 153681
GGRRVAR 125622
ALGNGKS 109876
LLGPGKT 107267
GRFVVEV 105507
TPHDFEV 88676
Time for index table init: 0h 4m 54s 821ms
Process prefiltering step 1 of 1
k-mer similarity threshold: 163
Starting prefiltering scores calculation (step 1 of 1)
Query db start 1 to 2531
Target db start 1 to 106052079
[=================================================================] 100.00% 2.53K 0s 910ms
31.980804 k-mers per position
170052 DB matches per sequence
0 overflows
0 queries produce too many hits (truncated result)
235 sequences passed prefiltering per query sequence
300 median result list length
0 sequences with 0 size result lists
Time for merging to pref_0: 0h 0m 0s 7ms
Time for processing: 0h 5m 20s 93ms
lcaalign /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/tmp_hsp1/10382565990596377146/pref_0 /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/first --sub-mat nucl:nucleotide.out,aa:blosum62.out -a 0 --alignment-mode 1 --alignment-output-mode 0 --wrapped-scoring 0 -e 1 --min-seq-id 0 --min-aln-len 0 --seq-id-mode 0 --alt-ali 0 -c 0 --cov-mode 0 --max-seq-len 65535 --comp-bias-corr 1 --max-rejected 5 --max-accept 30 --add-self-matches 0 --db-load-mode 0 --pca 1 --pcb 1.5 --score-bias 0 --realign 0 --realign-score-bias -0.2 --realign-max-seqs 2147483647 --gap-open nucl:5,aa:11 --gap-extend nucl:2,aa:1 --zdrop 40 --threads 112 --compressed 0 -v 3
Compute score only
Query database size: 2531 type: Aminoacid
Target database size: 106052079 type: Aminoacid
[=================================================================] 100.00% 2.53K 0s 737ms
Time for merging to first: 0h 0m 0s 7ms
59140 alignments calculated
54382 sequence pairs passed the thresholds (0.919547 of overall calculated)
21.486368 hits per query sequence
Time for processing: 0h 0m 7s 61ms
lca /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/first /tmp/tmp5y44n4h6/10505879959436434455/orfs_tax --blacklist '12908:unclassified sequences,28384:other sequences' --tax-lineage 0 --compressed 0 --threads 112 -v 3
Node name 'unclassified sequences' does not match to be blocked name 'RBG-16-66-30'
Node name 'other sequences' does not match to be blocked name 'B14-G1 sp003648675'
[=================================================================] 100.00% 2.53K 0s 28ms
Taxonomy for 0 out of 13195 entries not found
Time for merging to orfs_tax: 0h 0m 0s 16ms
Time for processing: 0h 0m 3s 758ms
mvdb /tmp/tmp5y44n4h6/10505879959436434455/tmp_taxonomy/5836584711541975065/first /tmp/tmp5y44n4h6/10505879959436434455/orfs_tax_aln -v 3
Time for processing: 0h 0m 0s 2ms
swapdb /tmp/tmp5y44n4h6/10505879959436434455/orfs_filter_h /tmp/tmp5y44n4h6/10505879959436434455/orfs_h_swapped --split-memory-limit 0 --threads 112 --compressed 0 -v 3
[=================================================================] 100.00% 2.53K 0s 12ms
Computing offsets.
[=================================================================] 100.00% 2.53K 0s 3ms
Reading results.
[=================================================================] 100.00% 2.53K 0s 5ms
Output database: /tmp/tmp5y44n4h6/10505879959436434455/orfs_h_swapped
[=================================================================] 100.00% 392 0s 105ms
Time for merging to orfs_h_swapped: 0h 0m 0s 3ms
Time for processing: 0h 0m 0s 177ms
aggregatetaxweights /home/dell/.cache/SemiBin/mmseqs2-GTDB/GTDB /tmp/tmp5y44n4h6/10505879959436434455/orfs_h_swapped /tmp/tmp5y44n4h6/10505879959436434455/orfs_tax /tmp/tmp5y44n4h6/10505879959436434455/orfs_tax_aln easy_single_sample_output/mmseqs_contig_annotation/mmseqs_contig_annotation --tax-lineage 1 --compressed 0 --threads 112 -v 3
[=================================================================] 100.00% 392 0s 26ms
Time for merging to mmseqs_contig_annotation: 0h 0m 0s 6ms
Time for processing: 0h 0m 0s 143ms
createtsv /tmp/tmp5y44n4h6/contig_DB easy_single_sample_output/mmseqs_contig_annotation/mmseqs_contig_annotation easy_single_sample_output/mmseqs_contig_annotation/taxonomyResult.tsv
MMseqs Version: 13.45111
First sequence as representative false
Target column 1
Add full header false
Sequence source 0
Database output false
Threads 112
Compressed 0
Verbosity 3
Time for merging to taxonomyResult.tsv: 0h 0m 0s 4ms
Time for processing: 0h 0m 0s 133ms
2022-08-25 20:48:02,164 - Training model and clustering.
2022-08-25 20:48:02,165 - Start training from one sample.
2022-08-25 20:48:02,272 - Training model...
0%| | 0/20 [00:00<?, ?it/s]2022-08-25 20:48:10,698 - Generate training data of 0:
2022-08-25 20:48:10,739 - Number of must link pair:163
2022-08-25 20:48:10,739 - Number of can not link pair:9394
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████| 20/20 [00:22<00:00, 1.14s/it]
2022-08-25 20:48:25,037 - Training finished.
2022-08-25 20:48:25,049 - Start binning.
2022-08-25 20:48:28,352 - Calculating depth matrix.
2022-08-25 20:48:28,366 - Edges:9962
2022-08-25 20:48:33,595 - Reclustering.
2022-08-25 20:48:45,490 - Binning finish.
If you find SemiBin useful, please cite:
Pan, S., Zhu, C., Zhao, XM. et al. A deep siamese neural network improves metagenome-assembled genomes in microbiome datasets across different environments. Nat Commun 13, 2326 (2022). https://doi.org/10.1038/s41467-022-29843-y.ok ,我有一个问题,不知道大家有没有解决办法,就是可不可以将这个数据库放到别的地方,然后semibin通过一个函数调用该位置的GTDB数据?希望本帖对大家有帮助!




















 747
747











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








