
【Title】Identification of exosomal microRNA panel as diagnostic and prognostic biomarker for small cell lung cancer
【Publication Journal】Biomarker Research
【Publication Time】2023 Sep
1.Paper collected 5 serum non-cancerous lung nodules (control) and 5 serum samples from patients with SCLC for exosomal miRNA array analysis.
2.Author obtained 51 candidate exosomal miRNAs, 31 exosomal miRNAs with at least 2-fold significant (FC > |2.0| and p-value < 0.05) expression differences, 20 miRNAs) with the greatest increase or decrease according to the expression rate regardless of the p value.
3.Author selected 25 miRNAs validated via qRT-PCR analysis using the 5 serum non-cancerous lung nodules (control) and 5 serum samples from patients with SCLC.
That is to say, author selected miRNAs that express the same expression as array and RTQPCR.
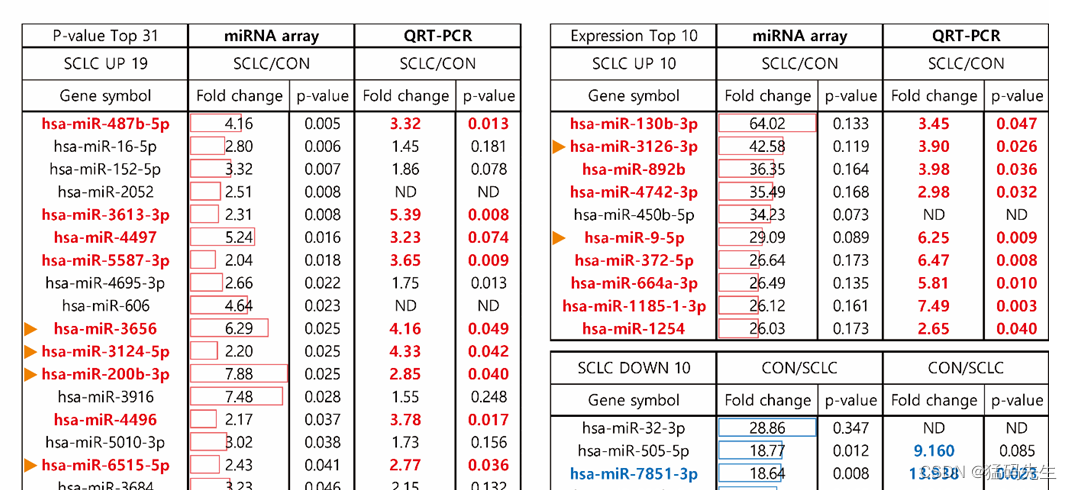
4.Next, 25 miRNAs were performed qRT-PCR in a larger cohort of “Test set” (50 healthy individuals and 76 SCLC patient serum derived exosomes). Author found 7 miRNAs with significantly different expression between the normal and SCLC among the 25 exosomal miRNAs.
Six miRNAs (miR-3656, miR-3124-5p, miR-200b-3p, miR-6515, miR-3126-3p and miR-9-5p) were up-regulated, and one miRNA (miR-92b-5p) was down-regulated in SCLC exosome
5.The expression of 7 miRNAs (predictors) and diagnosis result (outcome) of 126 biopsy samples (normal = 50, SCLC = 76) were used to develop the prediction models. R package Tidymodels (v0.1.4) was applied for modeling and machine learning.The samples were randomly selected and separated into training (94) and validation (32) set. All possible combinations of predictors with number of neighbors 3, 5, 7 were examined. The developed model was validated by diagnostic ability assessed via receiver operating characteristic (ROC) and area under the curve (AUC) calculation.
6. Among all 92 combinations, the highest area under the ROC curve (AUC) values and p-values were obtained when using 3-miRNA linear combination models (miR-200b-3p, miR-3124-5p and miR-92b-5p).
7. 3-miRNA were applied for distinguishing high-risk group and low-risk group in survival analysis.
8.miRNA target genes were derived using TargetScan8.0, miRDB and miRWalk online databases to identify functional parts of 3-miRNAs (miR-200b-3p, miR-3124-5p, miR-92b-5p).Overlapping genes from three online databases were considered as miRNA target genes. These target genes were applied for KEGG enrichment and GO term enrichment analysis.

























 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








