总结
把整个代码看完之后的总结,本代码中使用集成学习和5种重采样策略,使用回归器对数据进行预测。
- 所谓集成学习,就是使用多个回归器进行预测,对预测结果求平均值。
- 5种重采样策略+集成学习结合5种重采样策略= 10种
- 然后对一些超参数组组合进行训练,并将训练结果存储在excel文件中
- 评估指标:r2、mse、mcc、f1分数、每个箱子的mse
- 使用重采样策略提高酶的最佳催化温度(Topt)预测
- 把数据按照温度区间分成稀有域DR(大于72.2℃和小于20℃)和正常域DN(20℃-72.2℃)
引用包:
resreg: 回归的重采样策略
1. 计算氨基酸频率AA和取每个序列的生物体最适生长温度OGT
并对数据进行预处理:标准化,y值为Topt值。
aalist = list('ACDEFGHIKLMNPQRSTVWY')
def getAAC(seq):
aac = np.array([seq.count(x) for x in aalist])/len(seq)
return aac
data = pd.read_excel('data/sequence_ogt_topt.xlsx', index_col=0)
aac = np.array([getAAC(seq) for seq in data['sequence']])
ogt = data['ogt'].values.reshape((data.shape[0],1))
X = np.append(aac, ogt, axis=1)
sc = StandardScaler()
X = sc.fit_transform(X)
y = data['topt'].values2. 设置超参数与重采样策略
- l表示低极值,h表示高极值
- cl_vals、ch_values:表示设置稀有域和正常域的分隔温度(如上文中的72.2℃和20℃)。
通过网格搜索确定的超参数空间。
cl的值对应Topt的第10和20个百分位。最右侧的稀有域由None表示
ch的值对应Topt值的第90和80个百分位
- ks:插值选择的最近邻
- overs:WERCS方法中过采样的数量比例
- unders:WERCS方法中欠采样的数量比例
- sample_methods:采样的方法,对原始数据集重新采样后稀有域和正常域的个数重新分配
- size_methods:REBAGG算法的两种模式平衡模式和变化模式
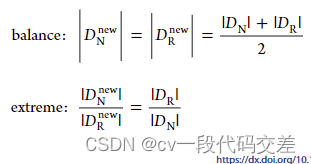
balance:从稀有域和正常域中随机抽取相等数量的样本S/2 .S为样本总数
variation:按照比例抽取样本,其中比例从集合中选取(1/3、2/5、1/2、3/5、2/3)
cl_vals = [25.0, 30.0, None]
ch_vals = [72.2, 60.0]
ks = [5, 10, 15]
deltas = [0.1, 0.5, 1.0]
overs = [0.5, 0.75]
unders = [0.5, 0.75]
sizes = [300, 600]
sample_methods = ['balance', 'extreme', 'average']
size_methods = ['balance', 'variation']
all_params = {}3. 超参数组合(网格搜索)
- 网格搜索:一种调参手段;穷举搜索:在所有候选的参数选择中,通过循环遍历,尝试每一种可能性,表现最好的参数就是最终的结果。其原理就像是在数组里找最大值。(为什么叫网格搜索?以有两个参数的模型为例,参数a有3种可能,参数b有4种可能,把所有可能性列出来,可以表示成一个3*4的表格,其中每个cell就是一个网格,循环过程就像是在每个网格里遍历、搜索,所以叫grid search)
- RO:随机过采样

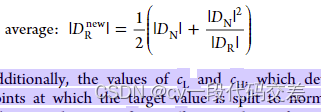
- SMOTER:用于回归的合成少数过采样技术:在DR中随机插值合成数据
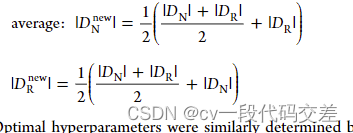
- GN:通过添加高斯噪声合成数据(采样方式与SMOTER相同)
- WERCS:基于加权相关性的组合策略通过相关值作为权重进行过采样和欠采样(复制和删除数据(没有正常域和稀有域)
计算每个值用于过采样和欠采样的概率进行采样。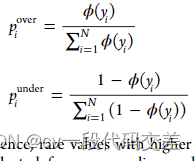
- WERCS-GN:向WERCS策略中选择的过采样值添加高斯噪声,也就是过采样使用合成数据。
- REBAGG-xx:各种采样策略与REBAGG算法相结合
product(*args):
接收若干个位置参数,转换成元组tuple形式
即几组参数随机组合
all_params['RO'] = list(itertools.product(cl_vals, ch_vals, sample_methods))
all_params['SMOTER'] = list(itertools.product(cl_vals, ch_vals, sample_methods, ks))
all_params['GN'] = list(itertools.product(cl_vals, ch_vals, sample_methods, deltas))
all_params['WERCS'] = list(itertools.product(cl_vals, ch_vals, overs, unders))
all_params['WERCS-GN'] = list(itertools.product(cl_vals, ch_vals, overs, unders, deltas))
all_params['REBAGG-RO'] = list(itertools.product(cl_vals, ch_vals, size_methods,
sizes))
all_params['REBAGG-SMOTER'] = list(itertools.product(cl_vals, ch_vals, size_methods,
sizes, ks))
all_params['REBAGG-GN'] = list(itertools.product(cl_vals, ch_vals, size_methods,
sizes, deltas))
all_params['REBAGG-WERCS'] = list(itertools.product(cl_vals, ch_vals, sizes, overs,
unders))
all_params['REBAGG-WERCS-GN'] = list(itertools.product(cl_vals, ch_vals, sizes, overs,
unders, deltas))
strategies = list(all_params.keys())4. 将每个策略应用于 Topt 预测并评估性能
bins:按照集合中的温度分成五个箱子
m:REBAGG 集成中的回归器数量
bins = [30, 50, 65, 85] # For splitting target values into bins
m = 100 # Number of regressors in REBAGG ensemble- 对于每一个策略和每一组超参数都计算(蒙特卡洛交叉验证 (MCCV) 循环):
- 划分训练集和测试集:测试集从5个箱子中分别选择70个,共350个,其余为训练集
- reg:回归模型
RO:
- resreg.sigmoid_relevance(y, cl, ch):使用sigmoid函数将数组y映射到相关值0-1
小于cl值的具有0.5的相关性,大于ch值的具有0.5的相关性(稀有域的全部0.5)
- random_oversample() 随机过采样
- oversample():随机过采样,并返回更大的数据集,即过采样后的数据集,随机复制样本添加到原样本中
def random_oversample(X, y, relevance, relevance_threshold=0.5, over='balance',
random_state=None):
X, y = np.asarray(X), np.squeeze(np.asarray(y))
relevance = np.squeeze(np.asarray(relevance))
[X_norm, y_norm, X_rare, y_rare] = split_domains(X, y, relevance, relevance_threshold)
# Determine size of rare domain after oversampling
if type(over)==float:
new_rare_size = int((1 + over) * len(y_rare))
elif over=='balance':
new_rare_size = len(y_norm)
elif over=='extreme':
new_rare_size = int(len(y_norm)**2 / len(y_rare))
elif over=='average':
new_rare_size = int((len(y_norm) + len(y_norm)**2 / len(y_rare)) / 2)
else:
raise ValueError('Incorrect value of over specified')
# Oversample rare domain
[X_rare_new, y_rare_new] = oversample(X_rare, y_rare, size=new_rare_size,
method='duplicate', random_state=random_state)
X_new = np.append(X_norm, X_rare_new, axis=0)
y_new = np.append(y_norm, y_rare_new, axis=0)
return [X_new, y_new]- SMOTER
smoter():
k:通过插值生成合成案例时使用的最近邻居数
- 根据relevance_threshold划分相关域和稀有域,小于的为正常域,大于为稀有域
- split_domains():根据阈值划分正常域和稀有域
- 欠采样后确定正常域的大小:三种采样方式
np.median(y):计算y的均值
np.where(y_rare < y_median):返回满足条件的坐标
low_indices、high_indices:比均值小和比均值大的坐标
对low_indices进行过采样,oversample()
elif method=='smoter':
if k is None:
raise ValueError("Must specify k if method is 'smoter'")
[X_more, y_more] = smoter_interpolate(X, y, k, size=moresize, nominal=nominal,
random_state=random_state)smoter_interpolate():通过在数据中的案例和随机选择的最近邻之间进行插值来生成新案例。对于正常特征,进行随机选择,而不是插值
- get_neighbors():返回X中每个案例的k个最近邻的索引
- dist():计算X矩阵每个元素之间的欧氏距离
- squareform():把距离矩阵下三角按列排成一个一维数组
- np.argsort():把数组中的元素从小到大排列,输出排序前的元素对应的索引
- neighbor_indices = np.array([row[1:k+1] for row in order]):间隔k取一个索引
def get_neighbors(X, k):
"""Return indices of k nearest neighbors for each case in X"""
X = np.asarray(X)
dist = pdist(X)
dist_mat = squareform(dist)
order = [np.argsort(row) for row in dist_mat]
neighbor_indices = np.array([row[1:k+1] for row in order])
return neighbor_indices- 生成数据:np.linalg.norm():求范数 元素的平方求和开根号。
def smoter_interpolate(X, y, k, size, nominal=None, random_state=None):
X, y = np.asarray(X), np.squeeze(np.asarray(y))
assert len(X)==len(y), 'X and y must be of the same length.'
neighbor_indices = get_neighbors(X, k) # Get indices of k nearest neighbors
np.random.seed(seed=random_state)
sample_indices = np.random.choice(range(len(y)), size, replace=True)
X_new, y_new = [], []
for i in sample_indices:
# Get case and nearest neighbor
X_case, y_case = X[i,:], y[i]
neighbor = np.random.choice(neighbor_indices[i,:])
X_neighbor, y_neighbor = X[neighbor, :], y[neighbor]
# Generate synthetic case by interpolation
rand = np.random.rand() * np.ones_like(X_case)
if nominal is not None:
rand = [np.random.choice([0,1]) if x in nominal else rand[x] \
for x in range(len(rand))] # Random selection for nominal features, rather than interpolation
rand = np.asarray(rand)
diff = (X_case - X_neighbor) * rand
X_new_case = X_neighbor + diff
d1 = np.linalg.norm(X_new_case - X_case)
d2 = np.linalg.norm(X_new_case - X_neighbor)
y_new_case = (d2 * y_case + d1 * y_neighbor) / (d2 + d1 + 1e-10) # Add 1e-10 to avoid division by zero
X_new.append(X_new_case)
y_new.append(y_new_case)
X_new = np.array(X_new)
y_new = np.array(y_new)
return [X_new, y_new]- Rebagg():
m:集成中模型的数量
s :为了拟合回归复制的样本数
fit():
- 复制m个回归器
- 随机挑选m个随机数1-2m之间
- sample():
- 采样方法和上面的采样方法一样
predict():
- 多个回归器预测求平均
def predict(self, X):
if not self._isfitted:
raise ValueError('Rebagg ensemble has not yet been fitted to training data.')
y_ensemble = np.array([reg.predict(X) for reg in self.fitted_regs])
y = np.mean(y_ensemble, axis=0)
self.pred_std = np.std(y_ensemble, axis=0)
assert len(y)==len(X), ('X and y do not have the same number of samples')
return ydef sample(self, X, y, relevance, relevance_threshold,
sample_method='random_oversample', size_method='balance', k=5, delta=0.1,
over=0.5, under=0.5, nominal=None, random_state=None):
X, y = np.asarray(X), np.squeeze(np.asarray(y))
relevance = np.squeeze(np.asarray(relevance))
assert len(y)==len(X), ('X and y do not have the same number of samples')
assert len(y)==len(relevance), 'y and relevance must be of the same length'
if type(self.s) == float:
size = int(self.s * X.shape[0])
elif type(self.s) == int:
size = self.s
np.random.seed(seed=random_state)
if sample_method in ('random_oversample', 'smoter', 'gaussian'):
# Split target values to rare and normal domains
X_norm_all, y_norm_all, X_rare_all, y_rare_all = split_domains(X, y,
relevance=relevance, relevance_threshold=relevance_threshold)
relevance_rare = relevance[np.where(relevance >= relevance_threshold)]
# Sample sizes
if size_method=='balance':
s_rare = int(size/2)
s_norm = size - s_rare
elif size_method=='variation':
p = np.random.choice([1/3, 2/5, 1/2, 2/5, 2/3])
s_rare = int(size * p)
s_norm = int(size - s_rare)
else:
raise ValueError('Wrong value of size_method specified')
# Sample rare data
if s_rare <= len(y_rare_all):
rare_indices = np.random.choice(range(len(y_rare_all)), s_rare,
replace=False) # No oversampling
X_rare, y_rare = X_rare_all[rare_indices,:], y_rare_all[rare_indices]
else:
if sample_method=='random_oversample':
rare_indices = np.random.choice(range(len(y_rare_all)), s_rare,
replace=True) # Duplicate samples
X_rare, y_rare = X_rare_all[rare_indices,:], y_rare_all[rare_indices]
elif sample_method=='smoter':
X_rare, y_rare = oversample(X_rare_all, y_rare_all, size=s_rare,
method='smoter', k=k,
relevance=relevance_rare, nominal=nominal,
random_state=random_state)
elif sample_method=='gaussian':
X_rare, y_rare = oversample(X_rare_all, y_rare_all, size=s_rare,
method='gaussian', delta=delta,
relevance=relevance_rare, nominal=nominal,
random_state=random_state)
# Sample normal data
norm_indices = np.random.choice(range(len(y_norm_all)), s_norm, replace=True)
X_norm, y_norm = X[norm_indices,:], y[norm_indices]
# Combine rare and normal samles
X_reg = np.append(X_rare, X_norm, axis=0)
y_reg = np.append(y_rare, y_norm, axis=0)
elif sample_method in ['wercs', 'wercs-gn']:
noise = True if sample_method=='wercs-gn' else False
X_reg, y_reg = wercs(X, y, relevance=relevance, over=over, under=under,
noise=noise, delta=delta, nominal=nominal,
random_state=random_state)
sample_indices = np.random.choice(range(len(y_reg)), size=size,
replace=(size>len(y_reg)))
X_reg, y_reg = X_reg[sample_indices,:], y_reg[sample_indices]
else:
raise ValueError("Wrong value of sample_method")
return [X_reg, y_reg]
def fit(self, X, y, relevance, relevance_threshold=0.5, sample_method='random_oversample',
size_method='balance', k=5, delta=0.1, over=0.5, under=0.5, nominal=None,
random_state=None):
regressors = [copy.deepcopy(self.base_reg) for i in range(self.m)]
np.random.seed(seed=random_state)
random_states = np.random.choice(range(2*self.m), size=self.m)
for i in range(self.m):
# Bootstrap samples
X_reg, y_reg = self.sample(X, y, relevance, relevance_threshold, sample_method,
size_method, k, delta, over, under, nominal,
random_states[i])
# Fit regressor to samples
regressors[i].fit(X_reg, y_reg)
self.fitted_regs = regressors
self._isfitted=True
rebagg = resreg.Rebagg(m=m, s=s, base_reg=reg)
rebagg.fit(X_train, y_train, relevance, relevance_threshold=0.5,
sample_method='random_oversample', size_method=size_method,
random_state=rrr)
- 评估验证集上的回归器性能:
计算r2、mse、mcc、真实值和预测值的相关性、f1分数、mse_bin
- mse_bins:bin_preformance():计算真实目标值的不同bin的预测性能
- 计算五个箱子的预测误差
r2 = r2_score(y_test, y_pred)
mse = mean_squared_error(y_test, y_pred)
mcc = resreg.matthews_corrcoef(y_test, y_pred, bins)
relevance_true = resreg.sigmoid_relevance(y_test, cl=None, ch=65)
relevance_pred = resreg.sigmoid_relevance(y_pred, cl=None, ch=65)
f1 = resreg.f1_score(y_test, y_pred, error_threshold=5,
relevance_true=relevance_true, relevance_pred=relevance_pred,
relevance_threshold=0.5, k=1e4)
mse_bins = resreg.bin_performance(y_test, y_pred, bins, metric='MSE')for strategy in strategies:
print(strategy)
params = all_params[strategy]
final_store = [] # For final results
for i_param in range(len(params)):
param = params[i_param]
r2_store, mse_store, mcc_store, f1_store = [], [], [], [] # Empty lists for storing results
mse_bins_store = []
# Monte Carlo cross validation (MCCV) loop
for rrr in range(50):
# Resample test set (uniform distribution)
train_indices, test_indices = resreg.uniform_test_split(X, y, bins=bins,
bin_test_size=70, verbose=False,
random_state=rrr)
X_train, y_train = X[train_indices,:], y[train_indices]
X_test, y_test = X[test_indices,:], y[test_indices]
# Unpack hyperparameters, resample training data, and fit regressors
reg = DecisionTreeRegressor(random_state=rrr) if 'REBAGG' in strategy else \
RandomForestRegressor(n_estimators=10, n_jobs=-1, random_state=rrr)
if strategy=='RO':
cl, ch, sample_method = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
X_train, y_train = resreg.random_oversample(X_train, y_train, relevance,
relevance_threshold=0.5, over=sample_method,
random_state=rrr)
reg.fit(X_train, y_train)
elif strategy=='SMOTER':
cl, ch, sample_method, k = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
X_train, y_train = resreg.smoter(X_train, y_train, relevance,
relevance_threshold=0.5, k=k, over=sample_method,
random_state=rrr)
reg.fit(X_train, y_train)
elif strategy=='GN':
cl, ch, sample_method, delta = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
X_train, y_train = resreg.gaussian_noise(X_train, y_train, relevance,
relevance_threshold=0.5, delta=delta, over=sample_method,
random_state=rrr)
reg.fit(X_train, y_train)
elif strategy=='WERCS':
cl, ch, over, under = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
X_train, y_train = resreg.wercs(X_train, y_train, relevance, over=over,
under=under, noise=False, random_state=rrr)
reg.fit(X_train, y_train)
elif strategy=='WERCS-GN':
cl, ch, over, under, delta = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
X_train, y_train = resreg.wercs(X_train, y_train, relevance, over=over,
under=under, noise=True, delta=delta, random_state=rrr)
reg.fit(X_train, y_train)
elif strategy=='REBAGG-RO':
cl, ch, size_method, s = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
rebagg = resreg.Rebagg(m=m, s=s, base_reg=reg)
rebagg.fit(X_train, y_train, relevance, relevance_threshold=0.5,
sample_method='random_oversample', size_method=size_method,
random_state=rrr)
elif strategy=='REBAGG-SMOTER':
cl, ch, size_method, s, k = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
rebagg = resreg.Rebagg(m=m, s=s, base_reg=reg)
rebagg.fit(X_train, y_train, relevance, relevance_threshold=0.5,
sample_method='smoter', size_method=size_method, k=k,
random_state=rrr)
elif strategy=='REBAGG-GN':
cl, ch, size_method, s, delta = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
rebagg = resreg.Rebagg(m=m, s=s, base_reg=reg)
rebagg.fit(X_train, y_train, relevance, relevance_threshold=0.5,
sample_method='gaussian', size_method=size_method, delta=delta,
random_state=rrr)
elif strategy=='REBAGG-WERCS':
cl, ch, s, over, under = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
rebagg = resreg.Rebagg(m=m, s=s, base_reg=reg)
rebagg.fit(X_train, y_train, relevance=relevance, sample_method='wercs',
over=over, under=under, random_state=rrr)
elif strategy=='REBAGG-WERCS-GN':
cl, ch, s, over, under, delta = param
relevance = resreg.sigmoid_relevance(y_train, cl=cl, ch=ch)
rebagg = resreg.Rebagg(m=m, s=s, base_reg=reg)
rebagg.fit(X_train, y_train, relevance=relevance, sample_method='wercs-gn',
over=over, under=under, delta=delta, random_state=rrr)
# Validate fitted regressors on uniform validation set
if 'REBAGG' in strategy:
y_pred = rebagg.predict(X_test)
else:
y_pred = reg.predict(X_test)
# Evaluate regressor performance on validation set
r2 = r2_score(y_test, y_pred)
mse = mean_squared_error(y_test, y_pred)
mcc = resreg.matthews_corrcoef(y_test, y_pred, bins)
relevance_true = resreg.sigmoid_relevance(y_test, cl=None, ch=65)
relevance_pred = resreg.sigmoid_relevance(y_pred, cl=None, ch=65)
f1 = resreg.f1_score(y_test, y_pred, error_threshold=5,
relevance_true=relevance_true, relevance_pred=relevance_pred,
relevance_threshold=0.5, k=1e4)
mse_bins = resreg.bin_performance(y_test, y_pred, bins, metric='MSE')
# Store performance results
r2_store.append(r2)
mse_store.append(mse)
mcc_store.append(mcc)
f1_store.append(f1)
mse_bins_store.append(mse_bins)
- 每一个参数策略计算50次,求平均
# Performance statistics
r2_mean, r2_std = np.mean(r2_store), np.std(r2_store)
mse_mean, mse_std = np.mean(mse_store), np.std(mse_store)
f1_mean, f1_std = np.mean(f1_store), np.std(f1_store)
mcc_mean, mcc_std = np.mean(mcc_store), np.std(mcc_store)
mse_bins_store = pd.DataFrame(mse_bins_store)
mse_bins_mean = np.mean(mse_bins_store, axis=0)
mse_bins_std = np.std(mse_bins_store, axis=0)- 存储所有参数及对应结果
# Write results to excel file
final_store = pd.DataFrame(final_store)
final_store.columns = ['params', 'r2', 'mse', 'f1', 'mcc'] + \
['mse_bin{0}'.format(x) for x in range(1,6)] + \
['r2_std', 'mse_std', 'f1_std', 'mcc_std'] + \
['mse_bin{0}_std'.format(x) for x in range(1,6)]
final_store.to_excel('results/{0}.xlsx'.format(strategy))























 2155
2155











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








