翻译笔记:https://zhuanlan.zhihu.com/p/21930884?refer=intelligentunit
作业讲解视频地址:http://www.mooc.ai/course/364/learn#lesson/2118
作业讲解
求损失Loss
多分类线性SVM: y=Wx y = W x
Hinge Loss(Max margin),margin=1:
注意:这个式子中只有错误的分类才会产生loss,即j=i 正确分类是没有loss的。
设置Delta:超参数 Δ Δ 应该被设置成什么值?需要通过交叉验证来求得吗?在绝大多数情况下该超参数设为 Δ=1.0 Δ = 1.0 都是安全的。
实现代码如下:
scores=X.dot(W)
margin=scores-scores[np.arange(num_train),y].reshape(num_train,1)+1
margin[np.arange(num_train),y]=0.0#正确的这一列不该计算,归零
margin=(margin>0)*margin
loss+=margin.sum()/num_train
loss+=0.5*reg*np.sum(W*W)#乘0.5,因为后面是平方,求导时有一个2可以与0.5相乘为1求梯度dW
- Loss计算公式:
- 梯度:
其中 1 1 是一个示性函数,如果括号中的条件为真,那么函数值为 ,如果为假,则函数值为 0 0 。虽然上述公式看起来复杂,但在代码实现的时候比较简单:只需要计算没有满足边界值的分类的数量(因此对损失函数产生了贡献),然后乘以 就是梯度了。注意,这个梯度只是对应正确分类的 W W 的行向量的梯度,那些 行的梯度是:
具体实现代码:
margin=(margin>0)*1
row_sum=np.sum(margin,axis=1)
margin[np.arange(num_train),y]=-row_sum
dW=X.T.dot(margin)/num_train+reg*W总体代码展示
打开svm.ipynb:
import random
import numpy as np
from cs231n.data_utils import load_CIFAR10
import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'数据加载和预处理
与之前KNN相同,加载数据后可以显示一部分样本
# Load the raw CIFAR-10 data.
cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
num_classes = len(classes)
samples_per_class = 7
for y, cls in enumerate(classes):
idxs = np.flatnonzero(y_train == y)
idxs = np.random.choice(idxs, samples_per_class, replace=False)
for i, idx in enumerate(idxs):
plt_idx = i * num_classes + y + 1
plt.subplot(samples_per_class, num_classes, plt_idx)
plt.imshow(X_train[idx].astype('uint8'))
plt.axis('off')
if i == 0:
plt.title(cls)
plt.show()将数据集分为训练集、交叉验证集和测试集,并从训练集中分出一个小的子集
num_training = 49000
num_validation = 1000
num_test = 1000
num_dev = 500
mask = range(num_training, num_training + num_validation)
X_val = X_train[mask]
y_val = y_train[mask]
mask = range(num_training)
X_train = X_train[mask]
y_train = y_train[mask]
# We will also make a development set, which is a small subset of
# the training set.
mask = np.random.choice(num_training, num_dev, replace=False)
X_dev = X_train[mask]
y_dev = y_train[mask]
# We use the first num_test points of the original test set as our
# test set.
mask = range(num_test)
X_test = X_test[mask]
y_test = y_test[mask]
print ('Train data shape: ', X_train.shape)
print ('Train labels shape: ', y_train.shape)
print ('Validation data shape: ', X_val.shape)
print ('Validation labels shape: ', y_val.shape)
print ('Test data shape: ', X_test.shape)
print ('Test labels shape: ', y_test.shape)OUTPUT:
Train data shape: (49000, 32, 32, 3)
Train labels shape: (49000,)
Validation data shape: (1000, 32, 32, 3)
Validation labels shape: (1000,)
Test data shape: (1000, 32, 32, 3)
Test labels shape: (1000,)
改变数据维度,将每一张图片用一行向量来表示:
# Preprocessing: reshape the image data into rows
X_train = np.reshape(X_train, (X_train.shape[0], -1))
X_val = np.reshape(X_val, (X_val.shape[0], -1))
X_test = np.reshape(X_test, (X_test.shape[0], -1))
X_dev = np.reshape(X_dev, (X_dev.shape[0], -1))
# As a sanity check, print out the shapes of the data
print ('Training data shape: ', X_train.shape)
print ('Validation data shape: ', X_val.shape)
print ('Test data shape: ', X_test.shape)
print ('dev data shape: ', X_dev.shape)OUTPUT:
Training data shape: (49000, 3072)
Validation data shape: (1000, 3072)
Test data shape: (1000, 3072)
dev data shape: (500, 3072)
Normalize data 基于训练集,算出图片的均值,并减去。
mean_image = np.mean(X_train, axis=0)
X_train -= mean_image
X_val -= mean_image
X_test -= mean_image
X_dev -= mean_image数据矩阵加上偏差列,方便之后的计算
X_train = np.hstack([X_train, np.ones((X_train.shape[0], 1))])
X_val = np.hstack([X_val, np.ones((X_val.shape[0], 1))])
X_test = np.hstack([X_test, np.ones((X_test.shape[0], 1))])
X_dev = np.hstack([X_dev, np.ones((X_dev.shape[0], 1))])
print (X_train.shape, X_val.shape, X_test.shape, X_dev.shape)OUTPUT:
(49000, 3073) (1000, 3073) (1000, 3073) (500, 3073)
SVM分类器的实现
打开cs231n\classifiers文件夹中的 linear_svm.py 完成SVM的实现代码。
损失函数和梯度
方法一:
def svm_loss_naive(W, X, y, reg):
dW = np.zeros(W.shape) # initialize the gradient as zero
# compute the loss and the gradient
num_classes = W.shape[1]
num_train = X.shape[0]
loss = 0.0
for i in range(num_train):
scores = X[i].dot(W)
correct_class_score = scores[y[i]]
for j in range(num_classes):
if j == y[i]:
continue
margin = scores[j] - correct_class_score + 1# delta=1
if margin > 0:#max(0,--)操作
loss += margin
dW[:,y[i]] += -X[i] #对应正确分类的梯度 (D,)
dW[:,j] += X[i] #对应不正确分类的梯度
# Right now the loss is a sum over all training examples, but we want it
# to be an average instead so we divide by num_train.
loss /= num_train
dW/=num_train
# Add regularization to the loss.
loss += 0.5 * reg * np.sum(W * W)
dW += reg*W
return loss, dW方法二 矢量化版:
def svm_loss_vectorized(W, X, y, reg):
loss = 0.0
dW = np.zeros(W.shape) # initialize the gradient as zero
num_train = X.shape[0]
scores = X.dot(W)
margin = scores-scores[np.arange(num_train),y].reshape(num_train,1)+1
margin[np.arange(num_train),y] = 0.0#正确的这一列不该计算,归零
margin = (margin > 0)*margin
loss += margin.sum()/num_train
loss += 0.5*reg*np.sum(W*W)
margin = (margin > 0)*1
row_sum = np.sum(margin,axis=1)
margin[np.arange(num_train),y] = -row_sum
dW = X.T.dot(margin)/num_train + reg*W
return loss, dW梯度检查
返回svm.ipynb:
loss, grad = svm_loss_naive(W, X_dev, y_dev, 0.0)
from cs231n.gradient_check import grad_check_sparse
f = lambda w: svm_loss_naive(w, X_dev, y_dev, 0.0)[0]
grad_numerical = grad_check_sparse(f, W, grad)
loss, grad = svm_loss_naive(W, X_dev, y_dev, 1e2)
f = lambda w: svm_loss_naive(w, X_dev, y_dev, 1e2)[0]
grad_numerical = grad_check_sparse(f, W, grad)OUTPUT:
numerical: -7.714121 analytic: -7.714121, relative error: 2.882459e-11
numerical: 6.498201 analytic: 6.498201, relative error: 1.616181e-11
numerical: -5.996422 analytic: -5.996422, relative error: 2.548793e-11
numerical: 18.051604 analytic: 18.051604, relative error: 2.654182e-11
numerical: -5.968500 analytic: -5.968500, relative error: 1.546593e-12
numerical: -9.307208 analytic: -9.307208, relative error: 3.240868e-11
numerical: 1.955191 analytic: 1.955191, relative error: 8.351098e-11
numerical: -1.443719 analytic: -1.443719, relative error: 2.464525e-10
numerical: 8.680369 analytic: 8.680369, relative error: 5.149589e-11
numerical: 3.160608 analytic: 3.160608, relative error: 9.645160e-11
numerical: 3.229153 analytic: 3.229153, relative error: 6.157453e-11
numerical: 19.706387 analytic: 19.706387, relative error: 1.384803e-11
numerical: 4.034088 analytic: 4.034088, relative error: 4.787692e-11
numerical: -12.270168 analytic: -12.270168, relative error: 1.866756e-11
numerical: -2.268660 analytic: -2.268660, relative error: 3.041155e-11
numerical: -0.057320 analytic: -0.057320, relative error: 6.521383e-09
numerical: -7.380878 analytic: -7.380878, relative error: 3.713221e-11
numerical: 11.643386 analytic: 11.643386, relative error: 4.090276e-11
numerical: 15.993779 analytic: 15.993779, relative error: 7.325546e-12
numerical: -4.908855 analytic: -4.908855, relative error: 7.685458e-11
其中,梯度检查函数为:
def grad_check_sparse(f, x, analytic_grad, num_checks=10, h=1e-5):
"""
sample a few random elements and only return numerical
in this dimensions.
"""
for i in range(num_checks):
ix = tuple([randrange(m) for m in x.shape])
oldval = x[ix]
x[ix] = oldval + h # increment by h
fxph = f(x) # evaluate f(x + h)
x[ix] = oldval - h # increment by h
fxmh = f(x) # evaluate f(x - h)
x[ix] = oldval # reset
grad_numerical = (fxph - fxmh) / (2 * h)
grad_analytic = analytic_grad[ix]
rel_error = abs(grad_numerical - grad_analytic) / (abs(grad_numerical) + abs(grad_analytic))
print ('numerical: %f analytic: %f, relative error: %e' % (grad_numerical, grad_analytic, rel_error))计算Loss和梯度
tic = time.time()
loss_naive, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.00001)
toc = time.time()
print ('Naive loss: %e computed in %fs' % (loss_naive, toc - tic))
from cs231n.classifiers.linear_svm import svm_loss_vectorized
tic = time.time()
loss_vectorized, _ = svm_loss_vectorized(W, X_dev, y_dev, 0.00001)
toc = time.time()
print ('Vectorized loss: %e computed in %fs' % (loss_vectorized, toc - tic))
# The losses should match but your vectorized implementation should be much faster.
print ('difference: %f' % (loss_naive - loss_vectorized))OUTPUT:
Naive loss: 9.169259e+00 computed in 0.243172s
Vectorized loss: 9.169259e+00 computed in 0.127999s
difference: -0.000000
tic = time.time()
_, grad_naive = svm_loss_naive(W, X_dev, y_dev, 0.00001)
toc = time.time()
print ('Naive loss and gradient: computed in %fs' % (toc - tic))
tic = time.time()
_, grad_vectorized = svm_loss_vectorized(W, X_dev, y_dev, 0.00001)
toc = time.time()
print ('Vectorized loss and gradient: computed in %fs' % (toc - tic))
# The loss is a single number, so it is easy to compare the values computed
# by the two implementations. The gradient on the other hand is a matrix, so
# we use the Frobenius norm to compare them.
difference = np.linalg.norm(grad_naive - grad_vectorized, ord='fro')
print ('difference: %f' % difference)OUTPUT:
Naive loss and gradient: computed in 0.279197s
Vectorized loss and gradient: computed in 0.008006s
difference: 0.000000
随机梯度下降
在linear_classifier.py 文件中实现SGD函数
更新权值和loss
def train(self, X, y, learning_rate=1e-3, reg=1e-5, num_iters=100,
batch_size=200, verbose=False):
"""
Train this linear classifier using stochastic gradient descent.
Inputs:
- X: A numpy array of shape (N, D) containing training data; there are N
training samples each of dimension D.
- y: A numpy array of shape (N,) containing training labels; y[i] = c
means that X[i] has label 0 <= c < C for C classes.
- learning_rate: (float) learning rate for optimization.
- reg: (float) regularization strength.
- num_iters: (integer) number of steps to take when optimizing
- batch_size: (integer) number of training examples to use at each step.
- verbose: (boolean) If true, print progress during optimization.
Outputs:
A list containing the value of the loss function at each training iteration.
"""
num_train, dim = X.shape
num_classes = np.max(y) + 1 # assume y takes values 0...K-1 where K is number of classes
if self.W is None:
# lazily initialize W
self.W = 0.001 * np.random.randn(dim, num_classes)
# Run stochastic gradient descent to optimize W
loss_history = []
for it in range(num_iters):
X_batch = None
y_batch = None
#########################################################################
# TODO: #
# Sample batch_size elements from the training data and their #
# corresponding labels to use in this round of gradient descent. #
# Store the data in X_batch and their corresponding labels in #
# y_batch; after sampling X_batch should have shape (dim, batch_size) #
# and y_batch should have shape (batch_size,) #
# #
# Hint: Use np.random.choice to generate indices. Sampling with #
# replacement is faster than sampling without replacement. #
#########################################################################
#子采样
mask = np.random.choice(num_train,batch_size,replace = False)#replace = False 没有重复
X_batch = X[mask]
y_batch = y[mask]
#########################################################################
# END OF YOUR CODE #
#########################################################################
# evaluate loss and gradient
loss, grad = self.loss(X_batch, y_batch, reg)
loss_history.append(loss)
# perform parameter update
#########################################################################
# TODO: #
# Update the weights using the gradient and the learning rate. #
#########################################################################
#梯度更新
self.W += -learning_rate * grad
#########################################################################
# END OF YOUR CODE #
#########################################################################
if verbose and it % 100 == 0:
print('iteration %d / %d: loss %f' % (it, num_iters, loss))
return loss_history返回svm.ipynb:
from cs231n.classifiers import LinearSVM
svm = LinearSVM()
tic = time.time()
loss_hist = svm.train(X_train, y_train, learning_rate=1e-7, reg=5e4,
num_iters=1500, verbose=True)
toc = time.time()
print ('That took %fs' % (toc - tic))OUTPUT:
iteration 0 / 1500: loss 783.675906
iteration 100 / 1500: loss 286.361632
iteration 200 / 1500: loss 107.706003
iteration 300 / 1500: loss 42.079802
iteration 400 / 1500: loss 18.928470
iteration 500 / 1500: loss 10.308477
iteration 600 / 1500: loss 7.065352
iteration 700 / 1500: loss 6.034884
iteration 800 / 1500: loss 5.524071
iteration 900 / 1500: loss 5.358315
iteration 1000 / 1500: loss 5.227159
iteration 1100 / 1500: loss 5.353792
iteration 1200 / 1500: loss 4.651034
iteration 1300 / 1500: loss 5.133628
iteration 1400 / 1500: loss 5.408001
That took 16.869082s
# A useful debugging strategy is to plot the loss as a function of
# iteration number:
plt.plot(loss_hist)
plt.xlabel('Iteration number')
plt.ylabel('Loss value')
plt.show()OUTPUT:
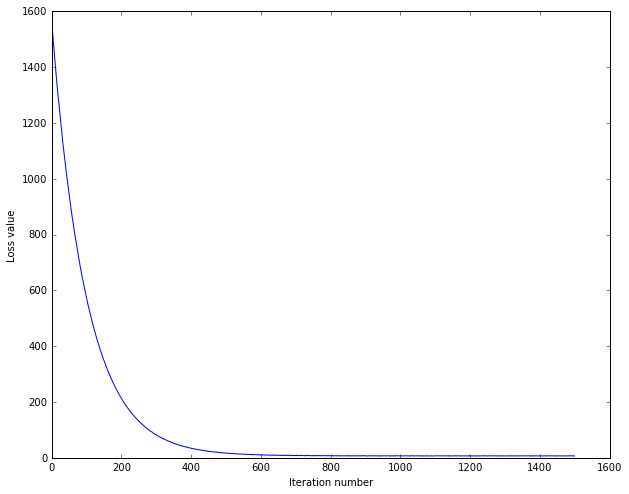
预测准确度
在linear_classifier.py 文件中实现predict函数
def predict(self, X):
"""
Use the trained weights of this linear classifier to predict labels for
data points.
Inputs:
- X: D x N array of training data. Each column is a D-dimensional point.
Returns:
- y_pred: Predicted labels for the data in X. y_pred is a 1-dimensional
array of length N, and each element is an integer giving the predicted
class.
"""
y_pred = np.zeros(X.shape[1])
###########################################################################
# TODO: #
# Implement this method. Store the predicted labels in y_pred. #
###########################################################################
score = X.dot(self.W)
index = np.zeros(X.shape[0])
index = np.argmax(score,axis = 1)
y_pred = index
###########################################################################
# END OF YOUR CODE #
###########################################################################
return y_pred得到预测label后,计算精度:
# Write the LinearSVM.predict function and evaluate the performance on both the
# training and validation set
y_train_pred = svm.predict(X_train)
print ('training accuracy: %f' % (np.mean(y_train == y_train_pred), ))
y_val_pred = svm.predict(X_val)
print ('validation accuracy: %f' % (np.mean(y_val == y_val_pred), ))利用交叉验证集调整超参数
# Use the validation set to tune hyperparameters (regularization strength and
# learning rate). You should experiment with different ranges for the learning
# rates and regularization strengths; if you are careful you should be able to
# get a classification accuracy of about 0.4 on the validation set.
learning_rates = [1e-7, 5e-5]
regularization_strengths = [5e4, 1e5]
# results is dictionary mapping tuples of the form
# (learning_rate, regularization_strength) to tuples of the form
# (training_accuracy, validation_accuracy). The accuracy is simply the fraction
# of data points that are correctly classified.
results = {}
best_val = -1 # The highest validation accuracy that we have seen so far.
best_svm = None # The LinearSVM object that achieved the highest validation rate.
################################################################################
# TODO: #
# Write code that chooses the best hyperparameters by tuning on the validation #
# set. For each combination of hyperparameters, train a linear SVM on the #
# training set, compute its accuracy on the training and validation sets, and #
# store these numbers in the results dictionary. In addition, store the best #
# validation accuracy in best_val and the LinearSVM object that achieves this #
# accuracy in best_svm. #
# #
# Hint: You should use a small value for num_iters as you develop your #
# validation code so that the SVMs don't take much time to train; once you are #
# confident that your validation code works, you should rerun the validation #
# code with a larger value for num_iters. #
################################################################################
iters = 2000 #100
for lr in learning_rates:
for rs in regularization_strengths:
svm = LinearSVM()
svm.train(X_train, y_train, learning_rate=lr, reg=rs, num_iters=iters)
y_train_pred = svm.predict(X_train)
acc_train = np.mean(y_train == y_train_pred)
y_val_pred = svm.predict(X_val)
acc_val = np.mean(y_val == y_val_pred)
results[(lr, rs)] = (acc_train, acc_val)
if best_val < acc_val:
best_val = acc_val
best_svm = svm
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out results.
for lr, reg in sorted(results):
train_accuracy, val_accuracy = results[(lr, reg)]
print ('lr %e reg %e train accuracy: %f val accuracy: %f' % (
lr, reg, train_accuracy, val_accuracy))
print ('best validation accuracy achieved during cross-validation: %f' % best_val)OUTPUT:
lr 1.000000e-07 reg 5.000000e+04 train accuracy: 0.370633 val accuracy: 0.379000
lr 1.000000e-07 reg 1.000000e+05 train accuracy: 0.358286 val accuracy: 0.366000
lr 5.000000e-05 reg 5.000000e+04 train accuracy: 0.100265 val accuracy: 0.087000
lr 5.000000e-05 reg 1.000000e+05 train accuracy: 0.100265 val accuracy: 0.087000
best validation accuracy achieved during cross-validation: 0.379000
可视化结果:
# Visualize the cross-validation results
import math
x_scatter = [math.log10(x[0]) for x in results]
y_scatter = [math.log10(x[1]) for x in results]
# plot training accuracy
marker_size = 100
colors = [results[x][0] for x in results]
plt.subplot(2, 1, 1)
plt.scatter(x_scatter, y_scatter, marker_size, c=colors)
plt.colorbar()
plt.xlabel('log learning rate')
plt.ylabel('log regularization strength')
plt.title('CIFAR-10 training accuracy')
# plot validation accuracy
colors = [results[x][1] for x in results] # default size of markers is 20
plt.subplot(2, 1, 2)
plt.scatter(x_scatter, y_scatter, marker_size, c=colors)
plt.colorbar()
plt.xlabel('log learning rate')
plt.ylabel('log regularization strength')
plt.title('CIFAR-10 validation accuracy')
plt.show()OUTPUT:
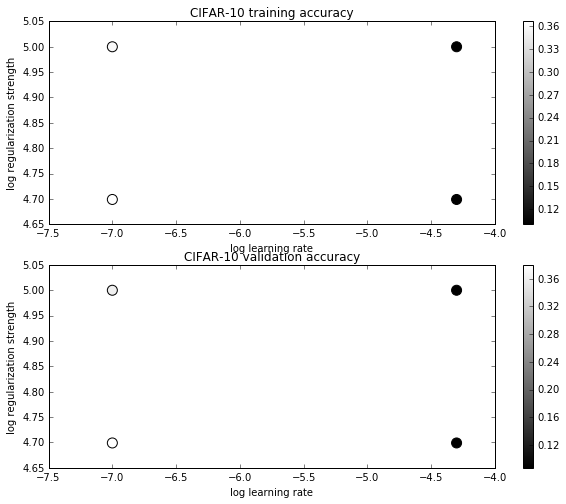
在测试集上验证
# Evaluate the best svm on test set
y_test_pred = best_svm.predict(X_test)
test_accuracy = np.mean(y_test == y_test_pred)
print ('linear SVM on raw pixels final test set accuracy: %f' % test_accuracy)OUTPUT: linear SVM on raw pixels final test set accuracy: 0.370000
# Visualize the learned weights for each class.
# Depending on your choice of learning rate and regularization strength, these may
# or may not be nice to look at.
w = best_svm.W[:-1,:] # strip out the bias
w = w.reshape(32, 32, 3, 10)
w_min, w_max = np.min(w), np.max(w)
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
for i in range(10):
plt.subplot(2, 5, i + 1)
# Rescale the weights to be between 0 and 255
wimg = 255.0 * (w[:, :, :, i].squeeze() - w_min) / (w_max - w_min)
plt.imshow(wimg.astype('uint8'))
plt.axis('off')
plt.title(classes[i])OUTPUT:









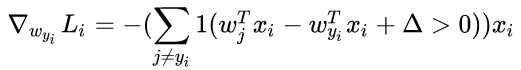
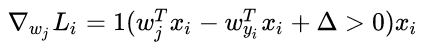














 1427
1427

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








