
阅读本文请先看 MIMIC-IV数据分析 - 使用Python进行心脏病案例分析
五、图 4:患者的意识水平是多少?
-
格拉斯哥昏迷量表 (GCS) 是一种意识测量方法。(神经功能的衡量标准)
-
范围从 3(最差,昏迷)到 15(最好,正常功能)。
-
-
它通常用于监测重症监护病房的患者
-
它由三个部分组成:眼睛反应;口头回应;运动反应。


import numpy as npimport pandas as pdimport matplotlib.pyplot as pltimport psycopg2schema_name = 'mimic'# 连接到MIMIC-IV数据库conn = psycopg2.connect(dbname='mimiciv', user='postgres', password='mimic',host='10.241.148.228', port=5432)query_schema = 'SET search_path to ' + schema_name + ';'# # # 设置查询语句# # # 我们选择从mimiciv_hosp.admissions表中提取hadm_id等于10006的行。# # # 在写sql代码时,最好先执行“set search_path to mimiciv" 随后的所有操作均不需要指明表格的位置;否则,任何操作都应该在表格名前面加前缀mimiciv# query1 = query_schema + 'SELECT subject_id, hadm_id, admittime, dischtime, admission_type FROM mimiciv_hosp.admissions'## # 运行查询并将结果分配给变量# admissions_pd = pd.read_sql_query(query1,conn)# admissions_pd.head()# print(admissions_pd.head())query = """SELECT de.stay_id, mimiciv_derived.datetime_diff(de.charttime, ie.intime, 'HOUR' ) AS HOURS, di.label, de.value, de.valuenum, de.valueuomFROM mimiciv_icu.chartevents deINNER join mimiciv_icu.d_items diON de.itemid = di.itemidINNER join mimiciv_icu.icustays ieON de.stay_id = ie.stay_idWHERE de.stay_id = 30121190ORDER BY charttime;"""ce = pd.read_sql_query(query, conn)# OPTION 2: load chartevents from a CSV file# ce = pd.read_csv('data/example_chartevents.csv', index_col='HOURSSINCEADMISSION')ce.head()# ce.label# print(ce.label)# Select just the heart rate rows using an indexd = ce[ce['label'].str.contains('Heart Rate')]# Display the first few rows of the GCS eye response datagcs = ce[ce['label'].str.contains('GCS - Eye Opening')]gcs_m = ce[ce['label'].str.contains('GCS - Motor Response')]gcs_v = ce[ce['label'].str.contains('GCS - Verbal Response')]print(gcs)r = ce[ce['label'].str.contains('Respiratory Rate')]# Prepare the size of the figureplt.figure(figsize=(12, 10))# # Set x equal to the timesx_hr = d['hours']## # Set y equal to the heart ratesy_hr = d['valuenum']## # Plot time against heart rateplt.plot(x_hr, y_hr)plt.plot(r['hours'],r['valuenum'],'k', markersize=6)# Add a text label to the y-axisplt.text(-20,155,'GCS - Eye Opening',fontsize=14)plt.text(-20,150,'GCS - Motor Response',fontsize=14)plt.text(-20,145,'GCS - Verbal Response',fontsize=14)# Iterate over list of GCS labels, plotting around 1 in 10 to avoid overlapfor i, txt in enumerate(gcs['value'].values):if np.mod(i,6)==0 and i < 65:plt.annotate(txt, (gcs['hours'].values[i],155),fontsize=14)for i, txt in enumerate(gcs_m['value'].values):if np.mod(i,6)==0 and i < 65:plt.annotate(txt, (gcs_m['hours'].values[i],150),fontsize=14)for i, txt in enumerate(gcs_v['value'].values):if np.mod(i,6)==0 and i < 65:plt.annotate(txt, (gcs_v['hours'].values[i],145),fontsize=14)plt.title('Vital signs and Glasgow Coma Scale over time from admission',fontsize=16)plt.xlabel('Time (hours)',fontsize=16)plt.ylabel('Heart rate or GCS',fontsize=16)plt.ylim(10,165)plt.show()
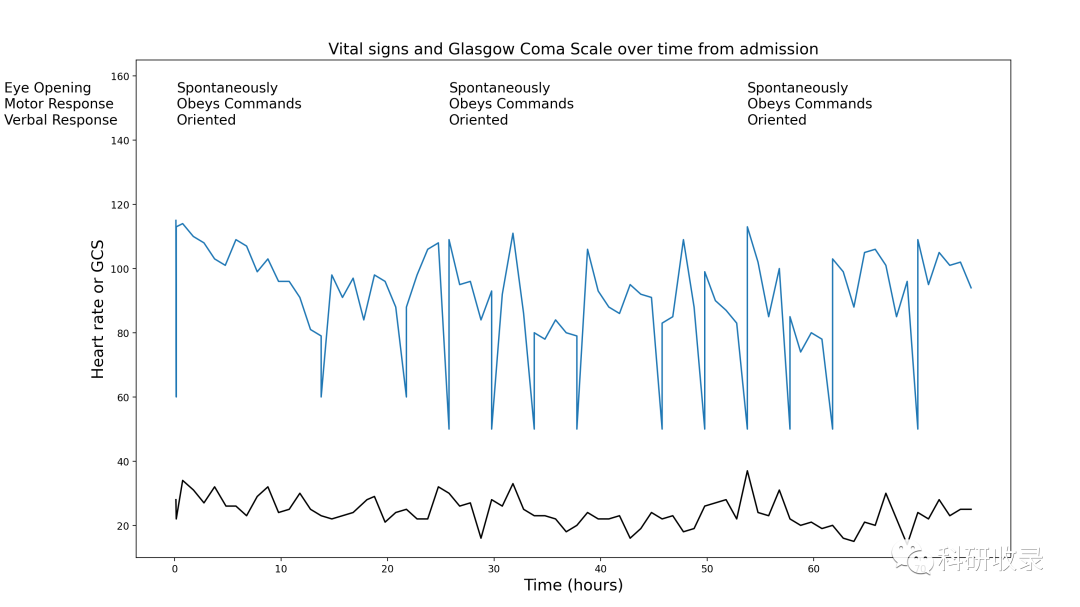
患者的意识随着时间的推移如何变化?
我们换个患者看看。WHERE de.stay_id = 36648401

关注【科研收录】回复"患者意识水平"获取以上代码
六、图 5:患者输出体液随时间变化情况
我们现在将查询 outputevents 输出事件表 该表包含有关患者输出的所有信息(尿液输出、引流、透析)。

import numpy as npimport pandas as pdimport matplotlib.pyplot as pltimport psycopg2schema_name = 'mimic'# 连接到MIMIC-IV数据库conn = psycopg2.connect(dbname='mimiciv', user='postgres', password='mimic',host='10.241.148.228', port=5432)query_schema = 'SET search_path to ' + schema_name + ';'# # # 设置查询语句# # # 我们选择从mimiciv_hosp.admissions表中提取hadm_id等于10006的行。# # # 在写sql代码时,最好先执行“set search_path to mimiciv" 随后的所有操作均不需要指明表格的位置;否则,任何操作都应该在表格名前面加前缀mimiciv# query1 = query_schema + 'SELECT subject_id, hadm_id, admittime, dischtime, admission_type FROM mimiciv_hosp.admissions'## # 运行查询并将结果分配给变量# admissions_pd = pd.read_sql_query(query1,conn)# admissions_pd.head()# print(admissions_pd.head())query = """SELECT de.stay_id, mimiciv_derived.datetime_diff(de.charttime, ie.intime, 'HOUR' ) AS HOURS, di.label, de.value, de.valueuomFROM mimiciv_icu.outputevents deINNER join mimiciv_icu.d_items diON de.itemid = di.itemidINNER join mimiciv_icu.icustays ieON de.stay_id = ie.stay_idWHERE de.subject_id = 10000980ORDER BY charttime;"""oe = pd.read_sql_query(query, conn)# OPTION 2: load chartevents from a CSV file# ce = pd.read_csv('data/example_chartevents.csv', index_col='HOURSSINCEADMISSION')oe.head()print(oe.head())



数据量有点少,我们换一个患者10001725






为了给该图提供必要的背景,图中最好包含患者的输入数据,即inputevents表中的数据
这能为确定患者的体液平衡(患者健康的关键指标)提供了必要的参考依据。


请注意,列标题不同:我们有“HOURS_START”和“HOURS_END”。这是因为输入是在固定的时间内管理的。
列举label字段有哪些值?(重复的不显示)


这里我们取一个数值比较多的患者以方便后面输入输出对比 。de.subject_id = 10001884

import numpy as npimport pandas as pdimport matplotlib.pyplot as pltimport psycopg2schema_name = 'mimic'# 连接到MIMIC-IV数据库conn = psycopg2.connect(dbname='mimiciv', user='postgres', password='mimic',host='10.241.148.228', port=5432)query_schema = 'SET search_path to ' + schema_name + ';'# # # 设置查询语句# # # 我们选择从mimiciv_hosp.admissions表中提取hadm_id等于10006的行。# # # 在写sql代码时,最好先执行“set search_path to mimiciv" 随后的所有操作均不需要指明表格的位置;否则,任何操作都应该在表格名前面加前缀mimiciv# query1 = query_schema + 'SELECT subject_id, hadm_id, admittime, dischtime, admission_type FROM mimiciv_hosp.admissions'## # 运行查询并将结果分配给变量# admissions_pd = pd.read_sql_query(query1,conn)# admissions_pd.head()# print(admissions_pd.head())query = """SELECT de.stay_id, mimiciv_derived.datetime_diff(de.starttime, ie.intime, 'HOUR' ) AS HOURS_START, mimiciv_derived.datetime_diff(de.endtime, ie.intime, 'HOUR' ) AS HOURS_END, di.label, de.amount, de.amountuom, de.rate, de.rateuomFROM mimiciv_icu.inputevents deINNER join mimiciv_icu.d_items diON de.itemid = di.itemidINNER join mimiciv_icu.icustays ieON de.stay_id = ie.stay_idWHERE de.subject_id = 10001884ORDER BY endtime;"""ie = pd.read_sql_query(query, conn)# OPTION 2: load chartevents from a CSV file# ce = pd.read_csv('data/example_chartevents.csv', index_col='HOURSSINCEADMISSION')ie.head()# print(ie.head())ie['label'].unique()# print(ie['label'].unique())ieml = ie[ie['amountuom'].str.contains('ml')]# print(ieml['hours_end'])# print(ieml['amount'])query2 = """SELECT de.stay_id, mimiciv_derived.datetime_diff(de.charttime, ie.intime, 'HOUR' ) AS HOURS, di.label, de.value, de.valueuomFROM mimiciv_icu.outputevents deINNER join mimiciv_icu.d_items diON de.itemid = di.itemidINNER join mimiciv_icu.icustays ieON de.stay_id = ie.stay_idWHERE de.subject_id = 10001884ORDER BY charttime;"""oe = pd.read_sql_query(query2, conn)plt.figure(figsize=(14, 10))# Plot the cumulative input against the cumulative outputplt.plot(ieml['hours_end'],ieml['amount'].cumsum()/1000,'go', markersize=8, label='Intake volume, L')plt.plot(oe['hours'],oe['value'].cumsum()/1000,'ro', markersize=8, label='Output volume, L')plt.title('Fluid balance over time',fontsize=16)plt.xlabel('Hours',fontsize=16)plt.ylabel('Volume (litres)',fontsize=16)# plt.ylim(0,38)plt.legend()plt.show()

如图所示,患者的摄入量往往高于其输出量(正如人们所期望的!),但在某些时期,它们几乎是一对一的。
关注【科研收录】回复"患者输出体液"获取以上代码
























 1314
1314











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








