
简 介
glmboost (Gradient Boosting with Component-wise Linear Models) 实现了优化一般风险函数的增强,利用组件(惩罚)最小二乘估计作为基础学习器,用于将各种广义线性和广义加性模型拟合到潜在的高维数据。演示了如何使用 glmboost 来拟合不同复杂性的可解释模型。作为一个例子,在整个教程中,使用ovarian数据集。
软件包安装
glmboost()来自软件包mboost,另外还需要安装几个常用的依赖包:
if(!require(mboost))
install.packages("mboost")
install.packages("mlr3")
install.packages("remotes")
remotes::install_github("mlr-org/mlr3extralearners")
install.packages("mlr3proba", repos = "https://mlr-org.r-universe.dev")数据读取
数据说明:
futime: survival or censoring time
fustat: censoring status
age: in years
resid.ds: residual disease present (1=no,2=yes)
rx: treatment group
ecog.ps: ECOG performance status (1 is better, see reference)
data(cancer, package="survival")
head(ovarian)## futime fustat age resid.ds rx ecog.ps
## 1 59 1 72.3315 2 1 1
## 2 115 1 74.4932 2 1 1
## 3 156 1 66.4658 2 1 2
## 4 421 0 53.3644 2 2 1
## 5 431 1 50.3397 2 1 1
## 6 448 0 56.4301 1 1 2实例操作
参数说明:
## S3 method for class 'matrix'
glmboost(x, y, center = TRUE, weights = NULL,
offset = NULL, family = Gaussian(),
na.action = na.pass, control = boost_control(), oobweights = NULL, ...)
Arguments
formula
a symbolic description of the model to be fit.
data
a data frame containing the variables in the model.
weights
an optional vector of weights to be used in the fitting process.
offset
a numeric vector to be used as offset (optional).
family
a Family object.
na.action
a function which indicates what should happen when the data contain NAs.
contrasts.arg
a list, whose entries are contrasts suitable for input to the contrasts replacement function and whose names are the names of columns of data containing factors. See model.matrix.default.
center
logical indicating of the predictor variables are centered before fitting.
control
a list of parameters controlling the algorithm. For more details see boost_control.
oobweights
an additional vector of out-of-bag weights, which is used for the out-of-bag risk (i.e., if boost_control(risk = "oobag")). This argument is also used internally by cvrisk.
x
design matrix. Sparse matrices of class Matrix can be used as well.
y
vector of responses.
...
additional arguments passed to mboost_fit; currently none.构建模型
根据说明ovarian数据集,构建模型:
library(mlr3extralearners)
library(mlr3)
library(mboost)
library(survival)
fm <- Surv(futime, fustat) ~ age + resid.ds + rx + ecog.ps
fit <- glmboost(fm, data = ovarian, family = CoxPH(), control = boost_control(mstop = 500),
center = FALSE)
summary(fit)
##
## Generalized Linear Models Fitted via Gradient Boosting
##
## Call:
## glmboost.formula(formula = fm, data = ovarian, family = CoxPH(), center = FALSE, control = boost_control(mstop = 500))
##
##
## Cox Partial Likelihood
##
## Loss function:
##
## Number of boosting iterations: mstop = 500
## Step size: 0.1
## Offset: 0
##
## Coefficients:
## age resid.ds rx
## 0.03607338 0.32120881 -0.14604217
## attr(,"offset")
## [1] 0
##
## Selection frequencies:
## age resid.ds rx
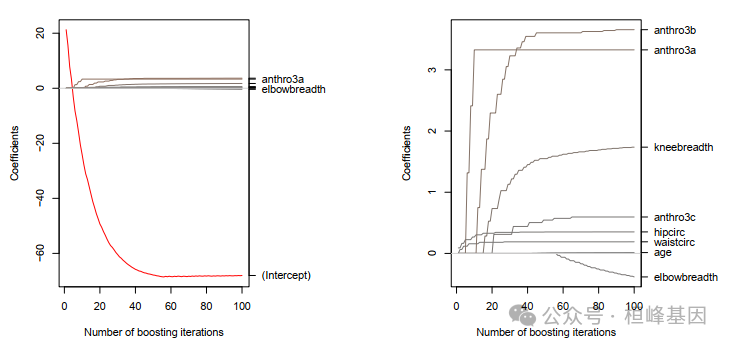
## 0.736 0.172 0.092plot(fit, main = "")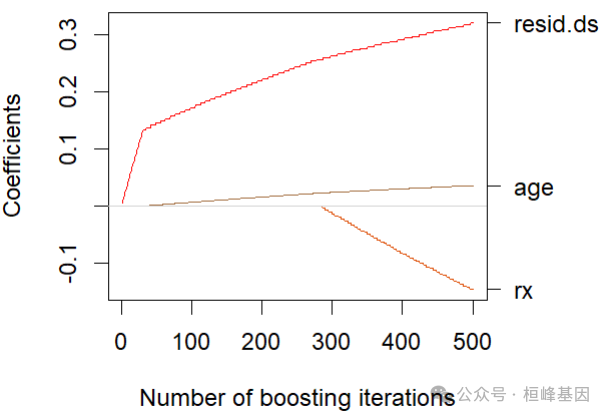
交叉验证
########
### 10-fold cross-validation
set.seed(123)
cv <- cv(model.weights(fit), type = "kfold")
cvm <- cvrisk(fit, folds = cv, papply = lapply)
mstop(cvm)## [1] 498plot(cvm)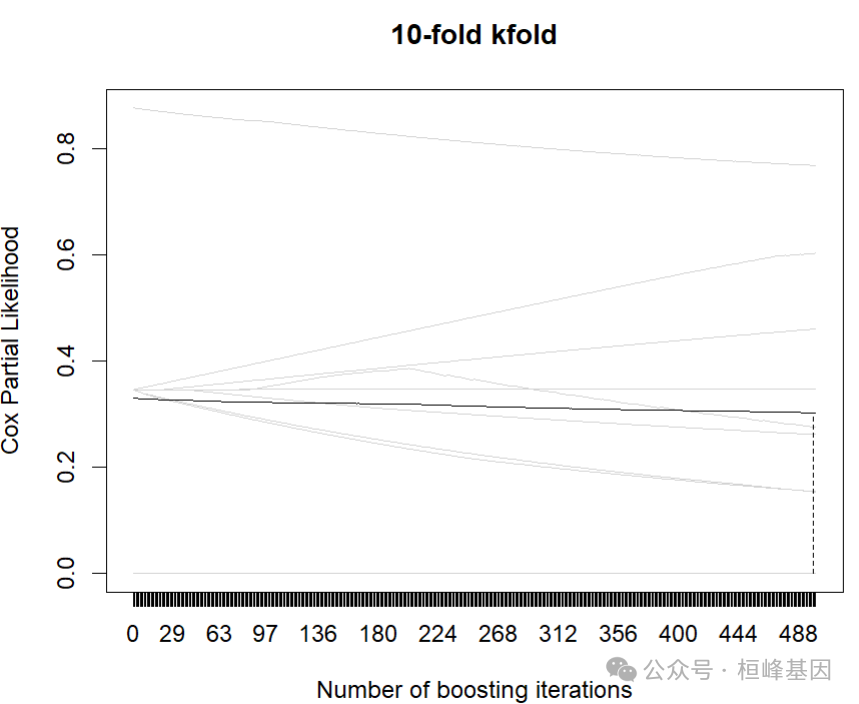
最优模型
OptFit <- glmboost(fm, data = ovarian, family = CoxPH(),
control=boost_control(mstop = 498),
center = FALSE)
plot(OptFit,main = "")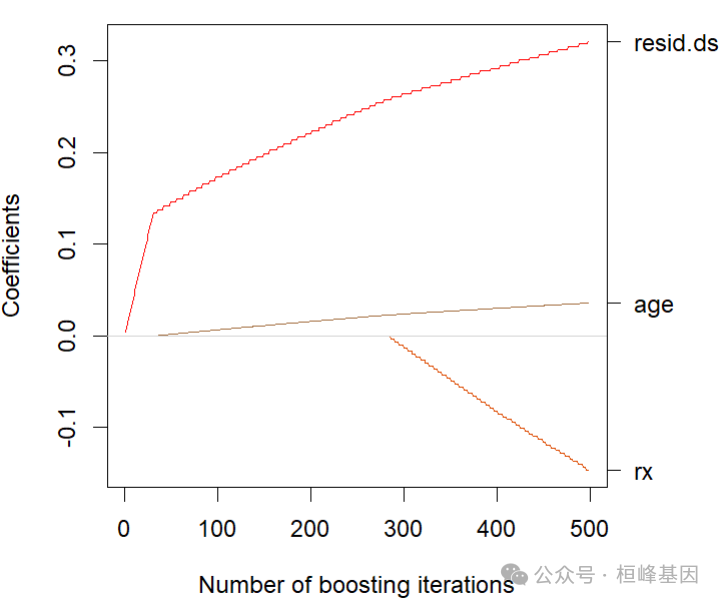
预测
pred <- predict(OptFit, newdata = ovarian)一致性
library(Hmisc)
rcorr.cens(pred, Surv(ovarian$futime, ovarian$fustat))
## C Index Dxy S.D. n missing
## 0.1880734 -0.6238532 0.1332940 26.0000000 0.0000000
## uncensored Relevant Pairs Concordant Uncertain
## 12.0000000 436.0000000 82.0000000 214.0000000生存分析
直接做生存分析即可:
### 生存分析
S1 <- survFit(OptFit)
S1
## $surv
## [,1]
## [1,] 0.9658296
## [2,] 0.9301417
## [3,] 0.8924592
## [4,] 0.8534872
## [5,] 0.8118936
## [6,] 0.7707809
## [7,] 0.7296796
## [8,] 0.6826778
## [9,] 0.6325667
## [10,] 0.5820787
## [11,] 0.5218996
## [12,] 0.4634418
##
## $time
## 1 2 3 22 23 24 25 5 7 8 10 11
## 59 115 156 268 329 353 365 431 464 475 563 638
##
## $n.event
## [1] 1 1 1 1 1 1 1 1 1 1 1 1
##
## attr(,"class")
## [1] "survFit"newdata <- ovarian[c(1, 3, 12), ]
S2 <- survFit(fit, newdata = newdata)
S2
## $surv
## 1 3 12
## [1,] 0.9261555 0.9398046 0.9786631
## [2,] 0.8523142 0.8786884 0.9560666
## [3,] 0.7779547 0.8161129 0.9318412
## [4,] 0.7049307 0.7535397 0.9063720
## [5,] 0.6312922 0.6891727 0.8786885
## [6,] 0.5628548 0.6280552 0.8507935
## [7,] 0.4987027 0.5694634 0.8223344
## [8,] 0.4305029 0.5055683 0.7890285
## [9,] 0.3637907 0.4411647 0.7525473
## [10,] 0.3027367 0.3802158 0.7146640
## [11,] 0.2378723 0.3128096 0.6678203
## [12,] 0.1829547 0.2529419 0.6203097
##
## $time
## 1 2 3 22 23 24 25 5 7 8 10 11
## 59 115 156 268 329 353 365 431 464 475 563 638
##
## $n.event
## [1] 1 1 1 1 1 1 1 1 1 1 1 1
##
## attr(,"class")
## [1] "survFit"par(mfrow = c(1, 2), mai = par("mai") * c(1, 1, 1, 2.5))
plot(S1, col = "red")
plot(S2)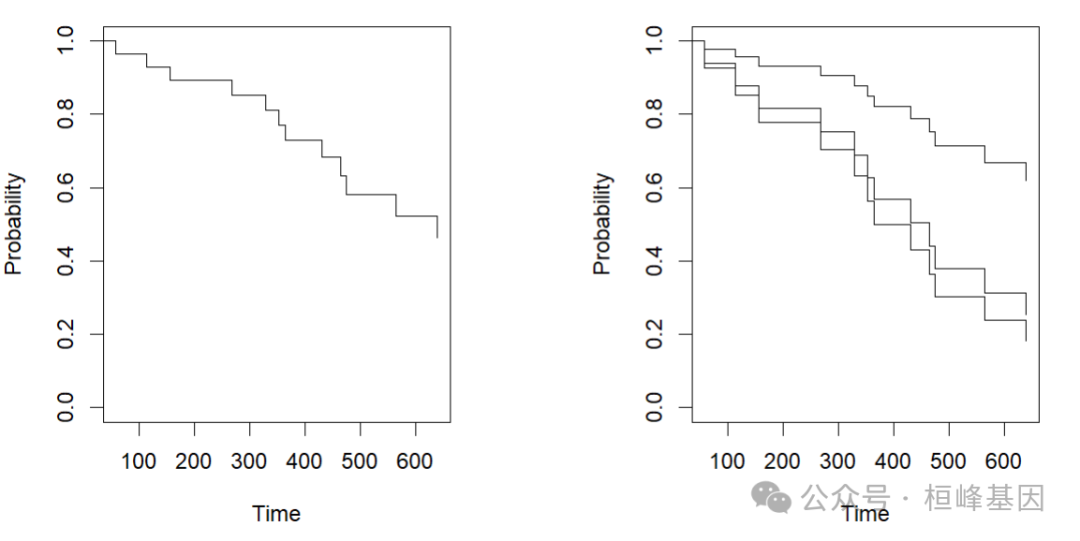
绘制ROC曲线
library(pROC)
roc <- roc(ovarian$fustat, pred, legacy.axes = T, print.auc = T, print.auc.y = 45)
roc$auc
## Area under the curve: 0.8274plot(roc, legacy.axes = T, col = "red", lwd = 2, main = "ROC curv")
text(0.2, 0.2, paste("AUC: ", round(roc$auc, 2)))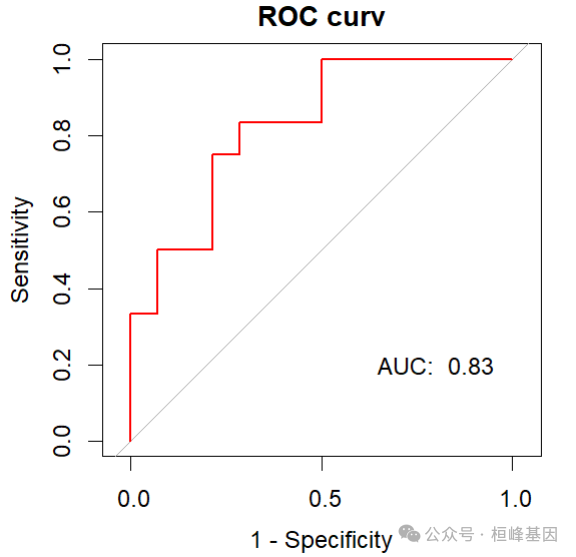
Reference
Peter Buehlmann and Bin Yu (2003), Boosting with the L2 loss: regression and classification. Journal of the American Statistical Association, 98, 324–339.
Peter Buehlmann (2006), Boosting for high-dimensional linear models. The Annals of Statistics, 34(2), 559–583.
Peter Buehlmann and Torsten Hothorn (2007), Boosting algorithms: regularization, prediction and model fitting. Statistical Science, 22(4), 477–505.
Torsten Hothorn, Peter Buehlmann, Thomas Kneib, Mattthias Schmid and Benjamin Hofner (2010), Model-based Boosting 2.0. Journal of Machine Learning Research, 11, 2109–2113.
Benjamin Hofner, Andreas Mayr, Nikolay Robinzonov and Matthias Schmid (2014). Model-based Boosting in R: A Hands-on Tutorial Using the R Package mboost. Computational Statistics, 29, 3–35.
机器学习构建临床预测模型
MachineLearning 2. 因子分析(Factor Analysis)
MachineLearning 3. 聚类分析(Cluster Analysis)
MachineLearning 4. 癌症诊断方法之 K-邻近算法(KNN)
MachineLearning 5. 癌症诊断和分子分型方法之支持向量机(SVM)
MachineLearning 6. 癌症诊断机器学习之分类树(Classification Trees)
MachineLearning 7. 癌症诊断机器学习之回归树(Regression Trees)
MachineLearning 8. 癌症诊断机器学习之随机森林(Random Forest)
MachineLearning 9. 癌症诊断机器学习之梯度提升算法(Gradient Boosting)
MachineLearning 10. 癌症诊断机器学习之神经网络(Neural network)
MachineLearning 11. 机器学习之随机森林生存分析(randomForestSRC)
MachineLearning 12. 机器学习之降维方法t-SNE及可视化(Rtsne)
MachineLearning 13. 机器学习之降维方法UMAP及可视化 (umap)
MachineLearning 14. 机器学习之集成分类器(AdaBoost)
MachineLearning 15. 机器学习之集成分类器(LogitBoost)
MachineLearning 16. 机器学习之梯度提升机(GBM)
MachineLearning 17. 机器学习之围绕中心点划分算法(PAM)
MachineLearning 18. 机器学习之贝叶斯分析类器(Naive Bayes)
MachineLearning 19. 机器学习之神经网络分类器(NNET)
MachineLearning 20. 机器学习之袋装分类回归树(Bagged CART)
MachineLearning 21. 机器学习之临床医学上的生存分析(xgboost)
MachineLearning 22. 机器学习之有监督主成分分析筛选基因(SuperPC)
MachineLearning 23. 机器学习之岭回归预测基因型和表型(Ridge)
MachineLearning 24. 机器学习之似然增强Cox比例风险模型筛选变量及预后估计(CoxBoost)
MachineLearning 25. 机器学习之支持向量机应用于生存分析(survivalsvm)
MachineLearning 26. 机器学习之弹性网络算法应用于生存分析(Enet)
MachineLearning 27. 机器学习之逐步Cox回归筛选变量(StepCox)
MachineLearning 28. 机器学习之偏最小二乘回归应用于生存分析(plsRcox)
MachineLearning 29. 机器学习之嵌套交叉验证(Nested CV)
MachineLearning 30. 机器学习之特征选择森林之神(Boruta)
MachineLearning 31. 机器学习之基于RNA-seq的基因特征筛选 (GeneSelectR)
MachineLearning 32. 机器学习之支持向量机递归特征消除的特征筛选 (mSVM-RFE)
MachineLearning 33. 机器学习之时间-事件预测与神经网络和Cox回归
MachineLearning 34. 机器学习之竞争风险生存分析的深度学习方法(DeepHit)
MachineLearning 35. 机器学习之Lasso+Cox回归筛选变量 (LassoCox)
MachineLearning 36. 机器学习之基于神经网络的Cox比例风险模型 (Deepsurv)
MachineLearning 37. 机器学习之倾斜随机生存森林 (obliqueRSF)
MachineLearning 38. 机器学习之基于最近收缩质心分类法的肿瘤亚型分类器 (pamr)
MachineLearning 39. 机器学习之基于条件随机森林的生存分析临床预测 (CForest)
MachineLearning 40. 机器学习之基于条件推理树的生存分析临床预测 (CTree)
MachineLearning 41. 机器学习之参数生存回归模型 (survreg)
MachineLearning 42. 机器学习之Akritas条件非参数生存估计 (akritas)
桓峰基因,铸造成功的您!
未来桓峰基因公众号将不间断的推出机器学习系列生信分析教程,
敬请期待!!
桓峰基因官网正式上线,请大家多多关注,还有很多不足之处,大家多多指正!http://www.kyohogene.com/
桓峰基因和投必得合作,文章润色优惠85折,需要文章润色的老师可以直接到网站输入领取桓峰基因专属优惠券码:KYOHOGENE,然后上传,付款时选择桓峰基因优惠券即可享受85折优惠哦!https://www.topeditsci.com/






















 8979
8979

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








