网上学习资料一大堆,但如果学到的知识不成体系,遇到问题时只是浅尝辄止,不再深入研究,那么很难做到真正的技术提升。
一个人可以走的很快,但一群人才能走的更远!不论你是正从事IT行业的老鸟或是对IT行业感兴趣的新人,都欢迎加入我们的的圈子(技术交流、学习资源、职场吐槽、大厂内推、面试辅导),让我们一起学习成长!
geom_edge_parallel(arrow=arrow(length=unit(1,"mm")),
aes(color=subtype_name),
end_cap=circle(7.5,"mm"),
alpha=0.5)+
geomtextpath::geom_textcontour(aes(x=x, y=y, z=z,color=after_stat(level)),
size=3, linetype=2,
linewidth=0.1, data=cont)+
geom_node_rect(aes(fill=padj, filter=type=="gene"))+
ggfx::with_outer_glow(geom_node_rect(aes(fill=padj, filter=!is.na(padj) & padj<0.05)),
colour="yellow", expand=2)+
geom_node_text(aes(label=converted_name), family="monocraft")+
scale_color_gradient2(low=scales::muted("blue"),
high=scales::muted("red"),
name="LFC")+
scale_edge_color_manual(values=viridis::viridis(11), name="Edge type")+
scale_fill_gradient(low="pink",high="steelblue") +
theme_void()
gg

### 将数值积分到`tbl_graph`
#### 将数值向量积分到`tbl_graph`
数值可以反映在节点或边表中,利用 或 函数。输入可以是命名向量,也可以是包含 id 和 value 列的 tibble。`node_numeric``edge_numeric`
vec <- 1
names(vec) <- c(“hsa:51343”)
new_g <- g |> mutate(num=node_numeric(vec))
new_g
#> # A tbl_graph: 134 nodes and 157 edges
#> #
#> # A directed acyclic multigraph with 40 components
#> #
#> # A tibble: 134 × 23
#> name type reaction graphics_name x y width
#>
#> 1 hsa:1029 gene CDKN2A, ARF,… 532 -218 46
#> 2 hsa:51343 gene FZR1, CDC20C… 981 -630 46
#> 3 hsa:4171 h… gene MCM2, BM28, … 553 -681 46
#> 4 hsa:23594 … gene ORC6, ORC6L.… 494 -681 46
#> 5 hsa:10393 … gene ANAPC10, APC… 981 -392 46
#> 6 hsa:10393 … gene ANAPC10, APC… 981 -613 46
#> # ℹ 128 more rows
#> # ℹ 16 more variables: height , fgcolor ,
#> # bgcolor , graphics_type , coords ,
#> # xmin , xmax , ymin , ymax ,
#> # orig.id , pathway_id , deseq2 ,
#> # padj , converted_name , lfc , num
#> #
#> # A tibble: 157 × 6
#> from to type subtype_name subtype_value pathway_id
#>
#> 1 118 39 GErel expression --> hsa04110
#> 2 50 61 PPrel inhibition --| hsa04110
#> 3 50 61 PPrel phosphorylation +p hsa04110
#> # ℹ 154 more rows
#### 将矩阵积分到`tbl_graph`
如果要在图形中反映表达式矩阵,则 和 函数可能很有用。通过指定基质和基因 ID,您可以将每个样品的数值分配给 . 分配由边连接的两个节点的总和,忽略组节点([Adnan 等人,2020]( ) 年)。`edge_matrix``node_matrix``tbl_graph``edge_matrix`
mat <- assay(vst(res))
new_g <- g |> edge_matrix(mat) |> node_matrix(mat)
new_g
#> # A tbl_graph: 134 nodes and 157 edges
#> #
#> # A directed acyclic multigraph with 40 components
#> #
#> # A tibble: 134 × 48
#> name type reaction graphics_name x y width
#>
#> 1 hsa:1029 gene CDKN2A, ARF,… 532 -218 46
#> 2 hsa:51343 gene FZR1, CDC20C… 981 -630 46
#> 3 hsa:4171 h… gene MCM2, BM28, … 553 -681 46
#> 4 hsa:23594 … gene ORC6, ORC6L.… 494 -681 46
#> 5 hsa:10393 … gene ANAPC10, APC… 981 -392 46
#> 6 hsa:10393 … gene ANAPC10, APC… 981 -613 46
#> # ℹ 128 more rows
#> # ℹ 41 more variables: height , fgcolor ,
#> # bgcolor , graphics_type , coords ,
#> # xmin , xmax , ymin , ymax ,
#> # orig.id , pathway_id , deseq2 ,
#> # padj , converted_name , lfc ,
#> # SRR14509882 , SRR14509883 , …
#> #
#> # A tibble: 157 × 34
#> from to type subtype_name subtype_value pathway_id
#>
#> 1 118 39 GErel expression --> hsa04110
#> 2 50 61 PPrel inhibition --| hsa04110
#> 3 50 61 PPrel phosphorylation +p hsa04110
#> # ℹ 154 more rows
#> # ℹ 28 more variables: from_nd , to_nd ,
#> # SRR14509882 , SRR14509883 ,
#> # SRR14509884 , SRR14509885 ,
#> # SRR14509886 , SRR14509887 ,
#> # SRR14509888 , SRR14509889 ,
#> # SRR14509890 , SRR14509891 , …
##### 边值
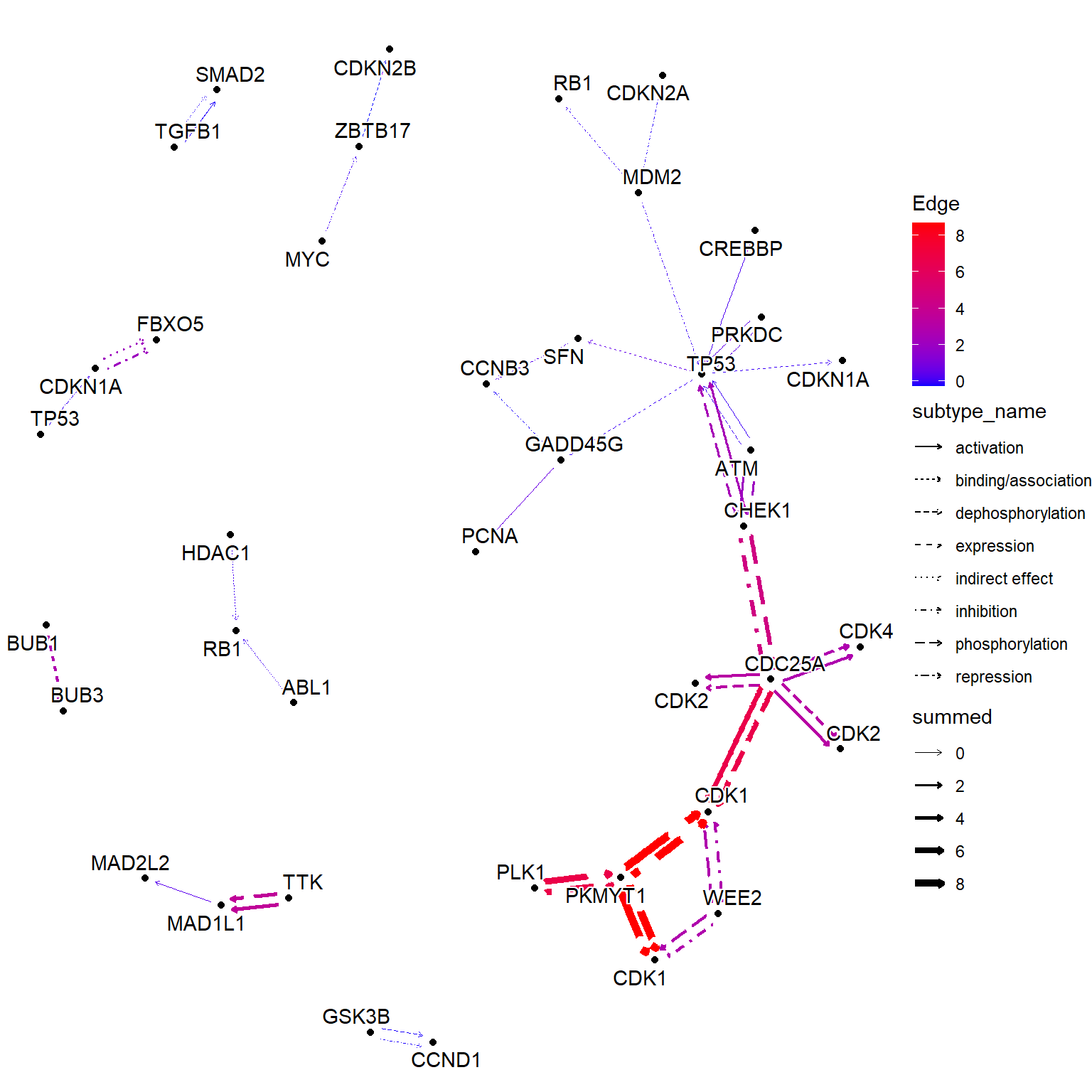
相同的效果可以通过 获得,使用命名数值向量作为输入。此函数根据节点值添加边值。以下示例显示了将 LFC 组合到边缘。这与 的行为不同。`edge_matrix``edge_numeric_sum``edge_numeric`
Numeric vector (name is SYMBOL)
vinflfc <- vinf$log2FoldChange |> setNames(row.names(vinf))
g |>
Use graphics_name to merge
mutate(grname=strsplit(graphics_name, “,”) |> vapply(“[”, 1, FUN.VALUE=“a”)) |>
activate(edges) |>
mutate(summed = edge_numeric_sum(vinflfc, name=“grname”)) |>
filter(!is.na(summed)) |>
activate(nodes) |>
mutate(x=NULL, y=NULL, deg=centrality_degree(mode=“all”)) |>
filter(deg>0) |>
ggraph(layout=“nicely”)+
geom_edge_parallel(aes(color=summed, width=summed,
linetype=subtype_name),
arrow=arrow(length=unit(1,“mm”)),
start_cap=circle(2,“mm”),
end_cap=circle(2,“mm”))+
geom_node_point(aes(fill=I(bgcolor)))+
geom_node_text(aes(label=grname,
filter=type==“gene”),
repel=TRUE, bg.colour=“white”)+
scale_edge_width(range=c(0.1,2))+
scale_edge_color_gradient(low=“blue”, high=“red”, name=“Edge”)+
theme_void()
### 可视化多重富集结果
您可以可视化多个富集分析的结果。与将函数与类一起使用类似,可以在函数中使用一个函数。通过向此功能提供对象,如果结果中存在可视化的通路,则通路内的基因信息可以反映在图中。在这个例子中,除了上面提到的尿路上皮细胞的变化外,还比较了肾近端肾小管上皮细胞的变化([Assetta等人,2016]( ))。`ggkegg``enrichResult``append_cp``mutate``enrichResult`
These are RDAs storing DEGs
load(“degListRPTEC.rda”)
load(“degURO.rda”)
library(org.Hs.eg.db);
library(clusterProfiler);
input_uro <- bitr(uroUp, ## DEGs in urothelial cells
fromType = “SYMBOL”,
toType = “ENTREZID”,
OrgDb = org.Hs.eg.db)
E
N
T
R
E
Z
I
D
i
n
p
u
t
r
p
t
e
c
<
−
b
i
t
r
(
g
l
s
ENTREZID input_rptec <- bitr(gls
ENTREZIDinputrptec<−bitr(glsday3_up_rptec, ## DEGs at 3 days post infection in RPTECs
fromType = “SYMBOL”,
toType = “ENTREZID”,
OrgDb = org.Hs.eg.db)$ENTREZID
ekuro <- enrichKEGG(gene = input_uro)
ekrptec <- enrichKEGG(gene = input_rptec)
g1 <- pathway(“hsa04110”) |> mutate(uro=append_cp(ekuro, how=“all”),
rptec=append_cp(ekrptec, how=“all”),
converted_name=convert_id(“hsa”))
ggraph(g1, layout=“manual”, x=x, y=y) +
geom_edge_parallel(width=0.5, arrow = arrow(length = unit(1, ‘mm’)),
start_cap = square(1, ‘cm’),
end_cap = square(1.5, ‘cm’), aes(color=subtype_name))+
geom_node_rect(aes(fill=uro, xmax=x, filter=type==“gene”))+
geom_node_rect(aes(fill=rptec, xmin=x, filter=type==“gene”))+
scale_fill_manual(values=c(“steelblue”,“tomato”), name=“urothelial|rptec”)+
ggfx::with_outer_glow(geom_node_text(aes(label=converted_name, filter=type!=“group”), size=2), colour=“white”, expand=1)+
theme_void()

我们可以按 组合多个图。`rawMap``patchwork`
library(patchwork)
comb <- rawMap(list(ekuro, ekrptec), fill_color=c(“tomato”,“tomato”), pid=“hsa04110”) +
rawMap(list(ekuro, ekrptec), fill_color=c(“tomato”,“tomato”),
pid=“hsa03460”)
comb

下面的示例将类似的反射应用于原始 KEGG 图谱,并突出显示在两种条件下都显示出统计学显着变化的基因,使用黄色外光,由 clusterProfiler 生成的组成,富集结果为 。`ggfx``dotplot``patchwork`
right <- (dotplot(ekuro) + ggtitle(“Urothelial”)) /
(dotplot(ekrptec) + ggtitle(“RPTECs”))
g1 <- pathway(“hsa03410”) |>
mutate(uro=append_cp(ekuro, how=“all”),
rptec=append_cp(ekrptec, how=“all”),
converted_name=convert_id(“hsa”))
gg <- ggraph(g1, layout=“manual”, x=x, y=y)+
ggfx::with_outer_glow(
geom_node_rect(aes(filter=uro&rptec),
color=“gold”, fill=“transparent”),
colour=“gold”, expand=5, sigma=10)+
geom_node_rect(aes(fill=uro, filter=type==“gene”))+
geom_node_rect(aes(fill=rptec, xmin=x, filter=type==“gene”)) +
overlay_raw_map(“hsa03410”, transparent_colors = c(“#cccccc”,“#FFFFFF”,“#BFBFFF”,“#BFFFBF”))+
scale_fill_manual(values=c(“steelblue”,“tomato”),
name=“urothelial|rptec”)+
theme_void()
gg2 <- gg + right + plot_layout(design="
AAAA###
AAAABBB
AAAABBB
AAAA###
"
)
gg2

#### 跨多个通路的多重富集分析结果
除了天然布局外,有时还可以在多个通路中显示有趣的基因,例如DEGs。在这里,我们使用[散点图]( )库来可视化跨多个途径的多个富集分析结果。
library(scatterpie)
Obtain enrichment analysis results
entrezid <- uroUp |>
clusterProfiler::bitr(“SYMBOL”,“ENTREZID”,org.Hs.eg.db)
cp <- clusterProfiler::enrichKEGG(entrezid$ENTREZID)
entrezid2 <- gls d a y 3 u p r p t e c ∣ > c l u s t e r P r o f i l e r : : b i t r ( " S Y M B O L " , " E N T R E Z I D " , o r g . H s . e g . d b ) c p 2 < − c l u s t e r P r o f i l e r : : e n r i c h K E G G ( e n t r e z i d 2 day3_up_rptec |> clusterProfiler::bitr("SYMBOL","ENTREZID",org.Hs.eg.db) cp2 <- clusterProfiler::enrichKEGG(entrezid2 day3uprptec∣>clusterProfiler::bitr("SYMBOL","ENTREZID",org.Hs.eg.db)cp2<−clusterProfiler::enrichKEGG(entrezid2ENTREZID)
Filter to interesting pathways
include <- (data.frame(cp) |> row.names())[c(1,3,4)]
pathways <- data.frame(cp)[include,“ID”]
pathways
#> [1] “hsa04110” “hsa03460” “hsa03440”
我们获得多个通路数据(该函数返回原生坐标,但我们忽略它们)。
g1 <- multi_pathway_native(pathways, row_num=1)
g2 <- g1 |> mutate(new_name=
ifelse(name==“undefined”,
paste0(name,““,pathway_id,””,orig.id),
name)) |>
convert(to_contracted, new_name, simplify=FALSE) |>
activate(nodes) |>
mutate(purrr::map_vec(.orig_data,function (x) x[1,] )) |>
mutate(pid1 = purrr::map(.orig_data,function (x) unique(x[“pathway_id”]) )) |>
mutate(hsa03440 = purrr:::map_lgl(pid1, function(x) “hsa03440” %in% x
p
a
t
h
w
a
y
i
d
)
,
h
s
a
04110
=
p
u
r
r
r
:
:
:
m
a
p
l
g
l
(
p
i
d
1
,
f
u
n
c
t
i
o
n
(
x
)
"
h
s
a
04110
"
pathway_id) , hsa04110 = purrr:::map_lgl(pid1, function(x) "hsa04110" %in% x
pathwayid),hsa04110=purrr:::maplgl(pid1,function(x)"hsa04110"pathway_id),
hsa03460 = purrr:::map_lgl(pid1, function(x) “hsa03460” %in% x$pathway_id))
nds <- g2 |> activate(nodes) |> data.frame()
eds <- g2 |> activate(edges) |> data.frame()
rmdup_eds <- eds[!duplicated(eds[,c(“from”,“to”,“subtype_name”)]),]
g2_2 <- tbl_graph(nodes=nds, edges=rmdup_eds)
g2_2 <- g2_2 |> activate(nodes) |>
mutate(
in_pathway_uro=append_cp(cp, pid=include,name=“new_name”),
x=NULL, y=NULL,
in_pathway_rptec=append_cp(cp2, pid=include,name = “new_name”),
id=convert_id(“hsa”,name = “new_name”)) |>
morph(to_subgraph, type!=“group”) |>
mutate(deg=centrality_degree(mode=“all”)) |>
unmorph() |>
filter(deg>0)
在这里,我们还将基于图的聚类结果分配给图,并缩放节点的大小,以便节点可以通过散点图可视化。
V(g2_2) w a l k t r a p < − i g r a p h : : w a l k t r a p . c o m m u n i t y ( g 2 2 ) walktrap <- igraph::walktrap.community(g2_2) walktrap<−igraph::walktrap.community(g22)membership
Scale the node size
sizeMin <- 0.1
sizeMax <- 0.3
rawMin <- min(V(g2_2)
d
e
g
)
r
a
w
M
a
x
<
−
m
a
x
(
V
(
g
2
2
)
deg) rawMax <- max(V(g2_2)
deg)rawMax<−max(V(g22)deg)
scf <- (sizeMax-sizeMin)/(rawMax-rawMin)
V(g2_2)
s
i
z
e
<
−
s
c
f
∗
V
(
g
2
2
)
size <- scf * V(g2_2)
size<−scf∗V(g22)deg + sizeMin - scf * rawMin
Make base graph
g3 <- ggraph(g2_2, layout=“nicely”)+
geom_edge_parallel(alpha=0.9,
arrow=arrow(length=unit(1,“mm”)),
aes(color=subtype_name),
start_cap=circle(3,“mm”),
end_cap=circle(8,“mm”))+
scale_edge_color_discrete(name=“Edge type”)
graphdata <- g3$data
最后,我们用于可视化。背景散点表示基因是否在通路中,前景表示基因是否在多个数据集中差异表达。我们突出显示了在两个数据集中通过金色差异表达的基因。`geom_scatterpie`
g4 <- g3+
ggforce::geom_mark_rect(aes(x=x, y=y, group=walktrap),color=“grey”)+
geom_scatterpie(aes(x=x, y=y, r=size+0.1),
color=“transparent”,
legend_name=“Pathway”,
data=graphdata,
cols=c(“hsa04110”, “hsa03440”,“hsa03460”)) +
geom_scatterpie(aes(x=x, y=y, r=size),
color=“transparent”,
data=graphdata, legend_name=“enrich”,
cols=c(“in_pathway_rptec”,“in_pathway_uro”))+
ggfx::with_outer_glow(geom_scatterpie(aes(x=x, y=y, r=size),
color=“transparent”,
data=graphdata[graphdataKaTeX parse error: Expected 'EOF', got '&' at position 18: …_pathway_rptec &̲ graphdatain_pathway_uro,],
cols=c(“in_pathway_rptec”,“in_pathway_uro”)), colour=“gold”, expand=3)+
geom_node_point(shape=19, size=3, aes(filter=!in_pathway_uro & !in_pathway_rptec & type!=“map”))+
geom_node_shadowtext(aes(label=id, y=y-0.5), size=3, family=“sans”, bg.colour=“white”, colour=“black”)+
theme_void()+coord_fixed()
g4

### 5.4 在KEGG图谱上投影基因调控网络
使用此软件包,可以将推断的网络(例如基因调控网络或由其他软件推断的 KO 网络)投射到 KEGG 图谱上。以下是使用 将 CBNplot 推断的通路内的 KO 网络子集投影到相应通路的参考图上的示例。当然,也可以投影使用其他方法创建的网络。`MicrobiomeProfiler`
library(dplyr)
library(igraph)
library(tidygraph)
library(CBNplot)
library(ggkegg)
library(MicrobiomeProfiler)
data(Rat_data)
ko.res <- enrichKO(Rat_data)
exp.dat <- matrix(abs(rnorm(910)), 91, 10) %>% magrittr::set_rownames(value=Rat_data) %>% magrittr::set_colnames(value=paste0(‘S’, seq_len(ncol(.))))
returnnet <- bngeneplot(ko.res, exp=exp.dat, pathNum=1, orgDb=NULL,returnNet = TRUE)
pg <- pathway(“ko00650”)
joined <- combine_with_bnlearn(pg, returnnet
s
t
r
,
r
e
t
u
r
n
n
e
t
str, returnnet
str,returnnetav)
绘制生成的地图。在此示例中,估计的强度首先用彩色边缘显示,然后参考图的边缘在其顶部以黑色绘制。此外,两个图形中包含的边缘都以黄色突出显示。`CBNplot`
Summarize duplicate edges including strength attribute
number <- joined |> activate(edges) |> data.frame() |> group_by(from,to) |>
summarise(n=n(), incstr=sum(!is.na(strength)))
Annotate them
joined <- joined |> activate(edges) |> full_join(number) |> mutate(both=n>1&incstr>0)
joined |>
activate(nodes) |>
filter(!is.na(type)) |>
mutate(convertKO=convert_id(“ko”)) |>
activate(edges) |>
ggraph(x=x, y=y) +
geom_edge_link0(width=0.5,aes(filter=!is.na(strength),
color=strength), linetype=1)+
ggfx::with_outer_glow(
geom_edge_link0(width=0.5,aes(filter=!is.na(strength) & both,
color=strength), linetype=1),
colour=“yellow”, sigma=1, expand=1)+
geom_edge_link0(width=0.1, aes(filter=is.na(strength)))+
scale_edge_color_gradient(low=“blue”,high=“red”)+
geom_node_rect(color=“black”, aes(fill=type))+
geom_node_text(aes(label=convertKO), size=2)+
geom_node_text(aes(label=ifelse(grepl(“:”, graphics_name), strsplit(graphics_name, “:”) |>
sapply(“[”,2) |> stringr::str_wrap(22), stringr::str_wrap(graphics_name, 22)),
filter=!is.na(type) & type==“map”), family=“serif”,
size=2, na.rm=TRUE)+
theme_void()

#### 5.4.1 投影到原始 KEGG 地图上
您可以直接将推断网络投影到原始 PATHWAY 地图上,这样可以直接比较您自己的数据集中精选数据库和推断网络的知识。
raws <- joined |>
ggraph(x=x, y=y) +
geom_edge_link(width=0.5,aes(filter=!is.na(strength),
color=strength),
linetype=1,
arrow=arrow(length=unit(1,“mm”),type=“closed”),
end_cap=circle(5,“mm”))+
scale_edge_color_gradient2()+
overlay_raw_map(transparent_colors = c(“#ffffff”))+
theme_void()
raws

### 5.5 分析单细胞转录组学中的簇标记基因
该软件包也可应用于单细胞分析。例如,考虑将簇之间的标记基因映射到 KEGG 通路上,并将它们与降维图一起绘制。在这里,我们使用包。我们进行基本面分析。`Seurat`
library(Seurat)
library(dplyr)
dir = “…/filtered_gene_bc_matrices/hg19”
pbmc.data <- Read10X(data.dir = dir)
pbmc <- CreateSeuratObject(counts = pbmc.data, project = “pbmc3k”,
min.cells=3, min.features=200)
pbmc <- NormalizeData(pbmc)
pbmc <- FindVariableFeatures(pbmc, selection.method = “vst”)
pbmc <- ScaleData(pbmc, features = row.names(pbmc))
pbmc <- RunPCA(pbmc, features = VariableFeatures(object = pbmc))
pbmc <- FindNeighbors(pbmc, dims = 1:10, verbose = FALSE)
pbmc <- FindClusters(pbmc, resolution = 0.5, verbose = FALSE)
markers <- FindAllMarkers(pbmc)
save(pbmc, markers, file=“…/sc_data.rda”)
To reduce file size, pre-calculated RDA will be loaded
load(“…/sc_data.rda”)
随后,我们绘制了PCA降维的结果。
其中,在本研究中,我们对簇 1 和 5 的标记基因进行了富集分析。
library(clusterProfiler)
Directly access slots in Seurat
pcas <- data.frame(
pbmc@reductions
p
c
a
@
c
e
l
l
.
e
m
b
e
d
d
i
n
g
s
[
,
1
]
,
p
b
m
c
@
r
e
d
u
c
t
i
o
n
s
pca@cell.embeddings[,1], pbmc@reductions
pca@cell.embeddings[,1],pbmc@reductionspca@cell.embeddings[,2],
pbmc@active.ident,
pbmc@meta.data$seurat_clusters) |>
colnames<-(c(“PC_1”,“PC_2”,“Cell”,“group”))
aa <- (pcas %>% group_by(Cell) %>%
mutate(meanX=mean(PC_1), meanY=mean(PC_2))) |>
select(Cell, meanX, meanY)
label <- aa[!duplicated(aa),]
dd <- ggplot(pcas)+
geom_point(aes(x=PC_1, y=PC_2, color=Cell))+
shadowtext::geom_shadowtext(x=label
m
e
a
n
X
,
y
=
l
a
b
e
l
meanX,y=label
meanX,y=labelmeanY,label=label$Cell, data=label,
bg.colour=“white”, colour=“black”)+
theme_minimal()+
theme(legend.position=“none”)
marker_1 <- clusterProfiler::bitr((markers |> filter(cluster==“1” & p_val_adj < 1e-50) |>
dplyr::select(gene))
g
e
n
e
,
f
r
o
m
T
y
p
e
=
"
S
Y
M
B
O
L
"
,
t
o
T
y
p
e
=
"
E
N
T
R
E
Z
I
D
"
,
O
r
g
D
b
=
o
r
g
.
H
s
.
e
g
.
d
b
)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb = org.Hs.eg.db)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb=org.Hs.eg.db)ENTREZID
marker_5 <- clusterProfiler::bitr((markers |> filter(cluster==“5” & p_val_adj < 1e-50) |>
dplyr::select(gene))
g
e
n
e
,
f
r
o
m
T
y
p
e
=
"
S
Y
M
B
O
L
"
,
t
o
T
y
p
e
=
"
E
N
T
R
E
Z
I
D
"
,
O
r
g
D
b
=
o
r
g
.
H
s
.
e
g
.
d
b
)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb = org.Hs.eg.db)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb=org.Hs.eg.db)ENTREZID
mk1_enrich <- enrichKEGG(marker_1)
mk5_enrich <- enrichKEGG(marker_5)
从中获取颜色信息,并使用 获取通路。在这里,我们选择了 ,节点根据降维图中的颜色着色,两个聚类中的标记都按指定的颜色 () 着色。这促进了通路信息(如KEGG)与单细胞分析数据之间的联系,从而能够创建直观且易于理解的视觉表示。`ggplot2``ggkegg``Osteoclast differentiation (hsa04380)``ggfx``tomato`
Make color map
built <- ggplot_build(dd)
d
a
t
a
[
[
1
]
]
c
o
l
s
<
−
b
u
i
l
t
data[[1]] cols <- built
data[[1]]cols<−builtcolour
names(cols) <- as.character(as.numeric(built$group)-1)
gr_cols <- cols[!duplicated(cols)]
g <- pathway(“hsa04380”) |> mutate(marker_1=append_cp(mk1_enrich),
marker_5=append_cp(mk5_enrich))
gg <- ggraph(g, layout=“manual”, x=x, y=y)+
geom_node_rect(aes(filter=marker_1&marker_5), fill=“tomato”)+ ## Marker 1 & 5
geom_node_rect(aes(filter=marker_1&!marker_5), fill=gr_cols[“1”])+ ## Marker 1
geom_node_rect(aes(filter=marker_5&!marker_1), fill=gr_cols[“5”])+ ## Marker 5
overlay_raw_map(“hsa04380”, transparent_colors = c(“#cccccc”,“#FFFFFF”,“#BFBFFF”,“#BFFFBF”))+
theme_void()
gg+dd+plot_layout(widths=c(0.6,0.4))

#### 5.5.1 组成多个通路的示例
我们可以在多种途径中检查标记基因,以更好地了解标记基因的作用。
library(clusterProfiler)
library(org.Hs.eg.db)
subset_lab <- label[label
C
e
l
l
d
d
<
−
g
g
p
l
o
t
(
p
c
a
s
)
+
g
g
f
x
:
:
w
i
t
h
o
u
t
e
r
g
l
o
w
(
g
e
o
m
n
o
d
e
p
o
i
n
t
(
s
i
z
e
=
1
,
a
e
s
(
x
=
P
C
1
,
y
=
P
C
2
,
f
i
l
t
e
r
=
g
r
o
u
p
=
=
"
1
"
,
c
o
l
o
r
=
g
r
o
u
p
)
)
,
c
o
l
o
u
r
=
"
t
o
m
a
t
o
"
,
e
x
p
a
n
d
=
3
)
+
g
g
f
x
:
:
w
i
t
h
o
u
t
e
r
g
l
o
w
(
g
e
o
m
n
o
d
e
p
o
i
n
t
(
s
i
z
e
=
1
,
a
e
s
(
x
=
P
C
1
,
y
=
P
C
2
,
f
i
l
t
e
r
=
g
r
o
u
p
=
=
"
5
"
,
c
o
l
o
r
=
g
r
o
u
p
)
)
,
c
o
l
o
u
r
=
"
t
o
m
a
t
o
"
,
e
x
p
a
n
d
=
3
)
+
g
g
f
x
:
:
w
i
t
h
o
u
t
e
r
g
l
o
w
(
g
e
o
m
n
o
d
e
p
o
i
n
t
(
s
i
z
e
=
1
,
a
e
s
(
x
=
P
C
1
,
y
=
P
C
2
,
f
i
l
t
e
r
=
g
r
o
u
p
=
=
"
4
"
,
c
o
l
o
r
=
g
r
o
u
p
)
)
,
c
o
l
o
u
r
=
"
g
o
l
d
"
,
e
x
p
a
n
d
=
3
)
+
g
g
f
x
:
:
w
i
t
h
o
u
t
e
r
g
l
o
w
(
g
e
o
m
n
o
d
e
p
o
i
n
t
(
s
i
z
e
=
1
,
a
e
s
(
x
=
P
C
1
,
y
=
P
C
2
,
f
i
l
t
e
r
=
g
r
o
u
p
=
=
"
6
"
,
c
o
l
o
r
=
g
r
o
u
p
)
)
,
c
o
l
o
u
r
=
"
g
o
l
d
"
,
e
x
p
a
n
d
=
3
)
+
s
h
a
d
o
w
t
e
x
t
:
:
g
e
o
m
s
h
a
d
o
w
t
e
x
t
(
x
=
s
u
b
s
e
t
l
a
b
Cell %in% c("1","4","5","6"),] dd <- ggplot(pcas) + ggfx::with_outer_glow(geom_node_point(size=1, aes(x=PC_1, y=PC_2, filter=group=="1", color=group)), colour="tomato", expand=3)+ ggfx::with_outer_glow(geom_node_point(size=1, aes(x=PC_1, y=PC_2, filter=group=="5", color=group)), colour="tomato", expand=3)+ ggfx::with_outer_glow(geom_node_point(size=1, aes(x=PC_1, y=PC_2, filter=group=="4", color=group)), colour="gold", expand=3)+ ggfx::with_outer_glow(geom_node_point(size=1, aes(x=PC_1, y=PC_2, filter=group=="6", color=group)), colour="gold", expand=3)+ shadowtext::geom_shadowtext(x=subset_lab
Celldd<−ggplot(pcas)+ggfx::withouterglow(geomnodepoint(size=1,aes(x=PC1,y=PC2,filter=group=="1",color=group)),colour="tomato",expand=3)+ggfx::withouterglow(geomnodepoint(size=1,aes(x=PC1,y=PC2,filter=group=="5",color=group)),colour="tomato",expand=3)+ggfx::withouterglow(geomnodepoint(size=1,aes(x=PC1,y=PC2,filter=group=="4",color=group)),colour="gold",expand=3)+ggfx::withouterglow(geomnodepoint(size=1,aes(x=PC1,y=PC2,filter=group=="6",color=group)),colour="gold",expand=3)+shadowtext::geomshadowtext(x=subsetlabmeanX,
y=subset_lab
m
e
a
n
Y
,
l
a
b
e
l
=
s
u
b
s
e
t
l
a
b
meanY, label=subset_lab
meanY,label=subsetlabCell,
data=subset_lab,
bg.colour=“white”, colour=“black”)+
theme_minimal()
marker_1 <- clusterProfiler::bitr((markers |> filter(cluster==“1” & p_val_adj < 1e-50) |>
dplyr::select(gene))
g
e
n
e
,
f
r
o
m
T
y
p
e
=
"
S
Y
M
B
O
L
"
,
t
o
T
y
p
e
=
"
E
N
T
R
E
Z
I
D
"
,
O
r
g
D
b
=
o
r
g
.
H
s
.
e
g
.
d
b
)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb = org.Hs.eg.db)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb=org.Hs.eg.db)ENTREZID
marker_5 <- clusterProfiler::bitr((markers |> filter(cluster==“5” & p_val_adj < 1e-50) |>
dplyr::select(gene))
g
e
n
e
,
f
r
o
m
T
y
p
e
=
"
S
Y
M
B
O
L
"
,
t
o
T
y
p
e
=
"
E
N
T
R
E
Z
I
D
"
,
O
r
g
D
b
=
o
r
g
.
H
s
.
e
g
.
d
b
)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb = org.Hs.eg.db)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb=org.Hs.eg.db)ENTREZID
marker_6 <- clusterProfiler::bitr((markers |> filter(cluster==“6” & p_val_adj < 1e-50) |>
dplyr::select(gene))
g
e
n
e
,
f
r
o
m
T
y
p
e
=
"
S
Y
M
B
O
L
"
,
t
o
T
y
p
e
=
"
E
N
T
R
E
Z
I
D
"
,
O
r
g
D
b
=
o
r
g
.
H
s
.
e
g
.
d
b
)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb = org.Hs.eg.db)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb=org.Hs.eg.db)ENTREZID
marker_4 <- clusterProfiler::bitr((markers |> filter(cluster==“4” & p_val_adj < 1e-50) |>
dplyr::select(gene))
g
e
n
e
,
f
r
o
m
T
y
p
e
=
"
S
Y
M
B
O
L
"
,
t
o
T
y
p
e
=
"
E
N
T
R
E
Z
I
D
"
,
O
r
g
D
b
=
o
r
g
.
H
s
.
e
g
.
d
b
)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb = org.Hs.eg.db)
gene,fromType="SYMBOL",toType="ENTREZID",OrgDb=org.Hs.eg.db)ENTREZID
mk1_enrich <- enrichKEGG(marker_1)
mk5_enrich <- enrichKEGG(marker_5)
mk6_enrich <- enrichKEGG(marker_6)
mk4_enrich <- enrichKEGG(marker_4)
g1 <- pathway(“hsa04612”) |> mutate(marker_4=append_cp(mk4_enrich),
marker_6=append_cp(mk6_enrich),
gene_name=convert_id(“hsa”))
gg1 <- ggraph(g1, layout=“manual”, x=x, y=y)+
overlay_raw_map(“hsa04612”, transparent_colors = c(“#FFFFFF”, “#BFBFFF”, “#BFFFBF”))+
ggfx::with_outer_glow(
geom_node_rect(aes(filter=marker_4&marker_6), fill=“white”),
colour=“gold”)+
ggfx::with_outer_glow(
geom_node_rect(aes(filter=marker_4&!marker_6), fill=“white”),
colour=gr_cols[“4”])+
ggfx::with_outer_glow(
geom_node_rect(aes(filter=marker_6&!marker_4), fill=“white”),
colour=gr_cols[“6”], expand=3)+
overlay_raw_map(“hsa04612”, transparent_colors = c(“#B3B3B3”, “#FFFFFF”, “#BFBFFF”, “#BFFFBF”))+
theme_void()
g2 <- pathway(“hsa04380”) |> mutate(marker_1=append_cp(mk1_enrich),
marker_5=append_cp(mk5_enrich))
gg2 <- ggraph(g2, layout=“manual”, x=x, y=y)+
ggfx::with_outer_glow(
geom_node_rect(aes(filter=marker_1&marker_5),
fill=“white”), ## Marker 1 & 5
colour=“tomato”)+
ggfx::with_outer_glow(
geom_node_rect(aes(filter=marker_1&!marker_5),
fill=“white”), ## Marker 1
colour=gr_cols[“1”])+
ggfx::with_outer_glow(
geom_node_rect(aes(filter=marker_5&!marker_1),
fill=“white”), ## Marker 5
colour=gr_cols[“5”])+
overlay_raw_map(“hsa04380”,
transparent_colors = c(“#cccccc”,“#FFFFFF”,“#BFBFFF”,“#BFFFBF”))+
theme_void()
left <- (gg2 + ggtitle(“Marker 1 and 5”)) /
(gg1 + ggtitle(“Marker 4 and 6”))
final <- left + dd + plot_layout(design="
AAAAA###
AAAAACCC
BBBBBCCC
BBBBB###
")
final

#### 5.5.2 原始地图上数值的条形图
对于它们在多个聚类中丰富的节点,我们可以绘制数值的条形图。引用的代码由 inscaven [提供]( )。
Assign lfc to graph
mark_4 <- clusterProfiler::bitr((markers |> filter(cluster==“4” & p_val_adj < 1e-50) |>
dplyr::select(gene))KaTeX parse error: Expected 'EOF', got '&' at position 128: …r(cluster=="6" &̲ p_val_adj < 1e…gene,fromType=“SYMBOL”,toType=“ENTREZID”,OrgDb = org.Hs.eg.db)
mark_4
l
f
c
<
−
m
a
r
k
e
r
s
[
m
a
r
k
e
r
s
lfc <- markers[markers
lfc<−markers[markerscluster==“4” & markers
g
e
n
e
gene %in% mark_4
geneSYMBOL,]
a
v
g
l
o
g
2
F
C
m
a
r
k
4
avg_log2FC mark_4
avglog2FCmark4hsa <- paste0(“hsa:”,mark_4
E
N
T
R
E
Z
I
D
)
m
a
r
k
6
ENTREZID) mark_6
ENTREZID)mark6lfc <- markers[markersKaTeX parse error: Expected 'EOF', got '&' at position 14: cluster=="6" &̲ markersgene %in% mark_4
S
Y
M
B
O
L
,
]
SYMBOL,]
SYMBOL,]avg_log2FC
mark_6
h
s
a
<
−
p
a
s
t
e
0
(
"
h
s
a
:
"
,
m
a
r
k
6
hsa <- paste0("hsa:",mark_6
hsa<−paste0("hsa:",mark6ENTREZID)
mk4lfc <- mark_4
l
f
c
n
a
m
e
s
(
m
k
4
l
f
c
)
<
−
m
a
r
k
4
lfc names(mk4lfc) <- mark_4
lfcnames(mk4lfc)<−mark4hsa
mk6lfc <- mark_6
l
f
c
n
a
m
e
s
(
m
k
6
l
f
c
)
<
−
m
a
r
k
6
lfc names(mk6lfc) <- mark_6
lfcnames(mk6lfc)<−mark6hsa
g1 <- g1 |> mutate(mk4lfc=node_numeric(mk4lfc),
mk6lfc=node_numeric(mk6lfc))
Make data frame containing necessary data from node
subset_df <- g1 |> activate(nodes) |> data.frame() |>
dplyr::filter(marker_4 & marker_6) |>
dplyr::select(orig.id, mk4lfc, mk6lfc, x, y, xmin, xmax, ymin, ymax) |>
tidyr::pivot_longer(cols=c(“mk4lfc”,“mk6lfc”))
Actually we dont need position list
pos_list <- list()
annot_list <- list()
for (i in subset_dfKaTeX parse error: Expected '}', got 'EOF' at end of input: …et_df[subset_dforig.id==i,]
ymin <- tmp
y
m
i
n
∣
>
u
n
i
q
u
e
(
)
y
m
a
x
<
−
t
m
p
ymin |> unique() ymax <- tmp
ymin∣>unique()ymax<−tmpymax |> unique()
xmin <- tmp
x
m
i
n
∣
>
u
n
i
q
u
e
(
)
x
m
a
x
<
−
t
m
p
xmin |> unique() xmax <- tmp
xmin∣>unique()xmax<−tmpxmax |> unique()
pos_list[[as.character(i)]] <- c(xmin, xmax,
ymin, ymax)
barp <- tmp |>
ggplot(aes(x=name, y=value, fill=name))+
geom_col(width=1)+
scale_fill_manual(values=c(gr_cols[“4”] |> as.character(),
gr_cols[“6”] |> as.character()))+
labs(x = NULL, y = NULL) +
coord_cartesian(expand = FALSE) +
theme(
legend.position = “none”,
panel.background = element_rect(fill = “transparent”, colour = NA),
line = element_blank(),
text = element_blank()
)
gbar <- ggplotGrob(barp)
panel_coords <- gbar
l
a
y
o
u
t
[
g
b
a
r
layout[gbar
layout[gbarlayout
n
a
m
e
=
=
"
p
a
n
e
l
"
,
]
g
b
a
r
m
o
d
<
−
g
b
a
r
[
p
a
n
e
l
c
o
o
r
d
s
name == "panel", ] gbar_mod <- gbar[panel_coords
name=="panel",]gbarmod<−gbar[panelcoordst:panel_coords
b
,
p
a
n
e
l
c
o
o
r
d
s
b, panel_coords
b,panelcoordsl:panel_coords$r]
annot_list[[as.character(i)]] <- annotation_custom(gbar_mod,
xmin=xmin, xmax=xmax,
ymin=ymin, ymax=ymax)
}
Make ggraph, annotate barplot, and overlay raw map.
graph_tmp <- ggraph(g1, layout=“manual”, x=x, y=y)+
geom_node_rect(aes(filter=marker_4&marker_6),
fill=“gold”)+
geom_node_rect(aes(filter=marker_4&!marker_6),
fill=gr_cols[“4”])+
geom_node_rect(aes(filter=marker_6&!marker_4),
fill=gr_cols[“6”])+
theme_void()
final_bar <- Reduce(“+”, annot_list, graph_tmp)+
overlay_raw_map(“hsa04612”,
transparent_colors = c(“#FFFFFF”,
“#BFBFFF”,
“#BFFFBF”))
final_bar
最后的话
最近很多小伙伴找我要Linux学习资料,于是我翻箱倒柜,整理了一些优质资源,涵盖视频、电子书、PPT等共享给大家!
资料预览
给大家整理的视频资料:
给大家整理的电子书资料:

如果本文对你有帮助,欢迎点赞、收藏、转发给朋友,让我有持续创作的动力!
网上学习资料一大堆,但如果学到的知识不成体系,遇到问题时只是浅尝辄止,不再深入研究,那么很难做到真正的技术提升。
一个人可以走的很快,但一群人才能走的更远!不论你是正从事IT行业的老鸟或是对IT行业感兴趣的新人,都欢迎加入我们的的圈子(技术交流、学习资源、职场吐槽、大厂内推、面试辅导),让我们一起学习成长!
marker_6&!marker_4),
fill=gr_cols[“6”])+
theme_void()
final_bar <- Reduce(“+”, annot_list, graph_tmp)+
overlay_raw_map(“hsa04612”,
transparent_colors = c(“#FFFFFF”,
“#BFBFFF”,
“#BFFFBF”))
final_bar
最后的话
最近很多小伙伴找我要Linux学习资料,于是我翻箱倒柜,整理了一些优质资源,涵盖视频、电子书、PPT等共享给大家!
资料预览
给大家整理的视频资料:
[外链图片转存中…(img-Tk2ANNfp-1715101976818)]
给大家整理的电子书资料:
[外链图片转存中…(img-YQBoM2Xc-1715101976818)]
如果本文对你有帮助,欢迎点赞、收藏、转发给朋友,让我有持续创作的动力!
网上学习资料一大堆,但如果学到的知识不成体系,遇到问题时只是浅尝辄止,不再深入研究,那么很难做到真正的技术提升。
一个人可以走的很快,但一群人才能走的更远!不论你是正从事IT行业的老鸟或是对IT行业感兴趣的新人,都欢迎加入我们的的圈子(技术交流、学习资源、职场吐槽、大厂内推、面试辅导),让我们一起学习成长!






















 1675
1675

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








