从零开始用snakemake搭建完整的甲基化生信分析流程
阅读文章后的收获:专业的snakemake编程技能;甲基化分析pipeline
作者生物信息学硕士,6年生信分析师从业经验,快速响应读者问题,提供技术支持,欢迎订阅专栏
前言
需求:在linux服务器上部署甲基化分析pipeline,并用snakemake管理分析流程,本篇为基础篇,之后会带来详细的专业流程搭建方法
一、snakemake简介
snakemake起源时间为2012年,开发之前市面上已经有很多开源的workflow engine如Biopipe,Galaxy等,snakemake开发的目的是为了解决一些之前已有的workflow engine的弊端,核心优势在于只使用git就可以在远程服务器上部署生信分析workflow,并以类似python语言syntax的形式展现。感兴趣可以阅读作者文献。

snakemake原文献链接:点击阅读
二、甲基化生物信息分析流程
甲基化生信分析的核心是依托测序技术仪器如illumina产生fastq数据文件,主要是读取甲基化位点也就是fastq文件中的碱基C,核心还是测序,示例如下:
@A00511:46:H5JG2DSXX:4:1101:29776:4523 1:N:0:CGCTCATT+CCTATCCT
TATGAGAGAATGTATCCCCGTGTGTGTATGTATATTTAGAATTTTAGAAAATATTAATTATGGTTTTTAAATAGAAAAAATATGTTTTTATAAATTGGTAGAGATTATTTTAATTATGGATAAGATTTTTGAAAGAGTTATTAATTAGTT
+
FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF:FFFFFFFFFFFFFFFFFFFFFFF:FFFFFFFFFFFFFF:FFFF:FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF:FFFFFFFFFFFF,FFFFFFFFFFFFFFFFFFF
后面的分析都以这个fastq作为input,利用一些已经开源的常用分析软件,Bismark map到甲基化参考基因组,DSS R包差异甲基化统计学分析,R绘制heatmap,volcano图
甲基化pipeline中文件处理比较频繁,用snakemake管理workflow方便易维护,下面我们就开始分布写代码并讨论
三、分布书写snakemake代码
下面开始按步骤书写代码
流程1:fastp质控fastq文件:
fastp可以输入1个fastq文件(单端测序),也可以输入2个fastq文件(双端测序)
rule fastp:
input:
"C10Ca-18R11313_R1.fastq.gz", #输入第1个fastq文件
"C10Ca-18R11313_R2.fastq.gz" #输入第2个fastq文件
output:
"C10Ca-18R11313_R1pre.fastq.gz", #输出第1个fastq文件
"C10Ca-18R11313_R2pre.fastq.gz" #输出第2个fastq文件
shell:
"""mkdir -p test
fastp -i {input[0]} -I {input[1]} -o {output[0]} -O {output[1]}"""
执行结果,无报错,相当于使用命令:
fastp -i C10Ca-18R11313_R1.fastq.gz -I C10Ca-18R11313_R2.fastq.gz -o C10Ca-18R11313_R1pre.fastq.gz -O C10Ca-18R11313_R2pre.fastq.gz
运行结果如下
Building DAG of jobs...
Using shell: /usr/bin/bash
Provided cores: 1
Rules claiming more threads will be scaled down.
Job counts:
count jobs
1 fastp
1
[Thu Oct 14 17:23:16 2021]
rule fastp:
input: C10Ca-18R11313_R1.fastq.gz, C10Ca-18R11313_R2.fastq.gz
output: C10Ca-18R11313_R1pre.fastq.gz, C10Ca-18R11313_R2pre.fastq.gz
jobid: 0
Read1 before filtering:
total reads: 98502
total bases: 14775300
Q20 bases: 14338154(97.0414%)
Q30 bases: 13494654(91.3325%)
Read1 after filtering:
total reads: 97889
total bases: 14081251
Q20 bases: 13702479(97.3101%)
Q30 bases: 12916027(91.725%)
Read2 before filtering:
total reads: 98502
total bases: 14775300
Q20 bases: 14128760(95.6242%)
Q30 bases: 13163659(89.0923%)
Read2 aftering filtering:
total reads: 97889
total bases: 14081358
Q20 bases: 13605816(96.6229%)
Q30 bases: 12720227(90.3338%)
Filtering result:
reads passed filter: 195778
reads failed due to low quality: 660
reads failed due to too many N: 4
reads failed due to too short: 562
reads with adapter trimmed: 12268
bases trimmed due to adapters: 721851
JSON report: fastp.json
HTML report: fastp.html
fastp -i C10Ca-18R11313_R1.fastq.gz -I C10Ca-18R11313_R2.fastq.gz -o C10Ca-18R11313_R1pre.fastq.gz -O C10Ca-18R11313_R2pre.fastq.gz
fastp v0.14.1, time used: 3 seconds
[Thu Oct 14 17:23:19 2021]
Finished job 0.
1 of 1 steps (100%) done
Complete log: /public/DUAN/methylation/.snakemake/log/2021-10-14T172316.902475.snakemake.log
流程2:Bismark软件比对,使用上面质控后的fastq文件:
值得注意的是output这一个rule里面没有,原因是Bismark这一步生成的是一个结果文件夹,包含结果文件,无法指定生成文件的名字,所以在这个rule里就省略了
rule Bismark:
input:
"C10Ca-18R11313_R1pre.fastq.gz",
"C10Ca-18R11313_R2pre.fastq.gz"
shell:
"""/public/DUAN/methylation/bismark_ref/bismark --bowtie2 --path_to_bowtie /public/DUAN/methylation/bismark_ref/bowtie2-2.3.4.2-linux-x86_64 -N 0 -L 20 --quiet --un --ambiguous --sam -o /public/DUAN/methylation /home/lvhongyi/lvhy/reference/human/ensembl_hg19/bismark_ref -1 {input[0]} -2 {input[1]}"""
运行结果如下:
C methylated in CpG context: 60.9%
C methylated in CHG context: 0.5%
C methylated in CHH context: 0.5%
C methylated in unknown context (CN or CHN): 0.1%
Bismark completed in 0d 0h 2m 15s
====================
Bismark run complete
====================
[Tue Oct 19 11:09:16 2021]
Finished job 0.
1 of 1 steps (100%) done
Complete log: /public/DUAN/methylation/.snakemake/log/2021-10-19T110700.899050.snakemake.log
流程3:deduplicate_bismark去除重复序列,使用上面比对生成的sam文件:
rule Bismark_dedup:
input:
"C10Ca-18R11313_R1pre_bismark_bt2_pe.sam"
shell:
"""/public/DUAN/methylation/deduplicate_bismark -p {input} --output_dir /public/DUAN/methylation"""
运行结果如下:
skipping header line: @SQ SN:chrX LN:155270560
skipping header line: @SQ SN:chrY LN:59373566
skipping header line: @SQ SN:chrMT LN:16569
skipping header line: @PG ID:Bismark VN:v0.20.0 CL:"bismark --bowtie2 --path_to_bowtie /public/DUAN/methylation/bismark_ref/bowtie2-2.3.4.2-linux-x86_64 -N 0 -L 20 --quiet --un --ambiguous --sam -o /public/DUAN/methylation /home/lvhongyi/lvhy/reference/human/ensembl_hg19/bismark_ref -1 C10Ca-18R11313_R1pre.fastq.gz -2 C10Ca-18R11313_R2pre.fastq.gz"
Total number of alignments analysed in C10Ca-18R11313_R1pre_bismark_bt2_pe.sam: 84423
Total number duplicated alignments removed: 84052 (99.56%)
Duplicated alignments were found at: 142 different position(s)
Total count of deduplicated leftover sequences: 371 (0.44% of total)
[Tue Oct 19 11:14:38 2021]
Finished job 0.
1 of 1 steps (100%) done
Complete log: /public/DUAN/methylation/.snakemake/log/2021-10-19T111433.699220.snakemake.log
流程4:使用我修改过的封装好的python文件,从sam文件提取cov文件:
rule extract_CpG:
input:
"C10Ca-18R11313_R1pre_bismark_bt2_pe.deduplicated.sam"
shell:
"""python /public/DUAN/DMR/extract_CpG_data.py -i {input} -o /public/DUAN/methylation/C10Ca-18R11313.cov"""
运行结果如下:
/public/DUAN/DMR/extract_CpG_data.py:51: DeprecationWarning: 'U' mode is deprecated
f = open(filename, 'rU')
{}
[Tue Oct 19 11:22:00 2021]
Finished job 0.
1 of 1 steps (100%) done
生成的cov文件格式如下:
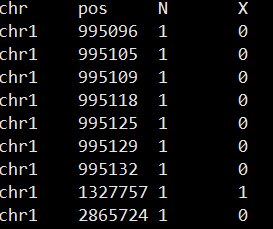
后续就可以用DSS读入cov文件进行分析组间甲基化差异啦






















 445
445











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










