对于靶向基因沉默,RNA干扰(RNAi)具有易于操作、快速结果、高效率、广泛适用于各种细胞类型的多重优势。其具有瞬转效应和剂量依赖效应,这点与小分子非常相似。然而,目前的siRNA试剂的特异性和基因沉默效率不是很稳定,阻碍了它们作为药物发现和基因研究工具的应用。

可靠表型的特异性基因沉默工具
siRNA pools(siPOOLs)是一款经过优化设计的、高复杂性的、包含30条siRNA的混合物,经证明可有效消除脱靶效应,提高了结果的可靠性(Hannus et al., 2014)。 Pack Hunter (pooling) 方法通过将当个siRNA的浓度稀释到刺激表型的阈值以下来对抗单个siRNA的脱靶。 借助专有的设计算法,siPOOLs中的siRNA序列经过优化,以实现最大的转录本覆盖率,高效杂交并对旁系同源基因进行过滤,从而实现高效和特异性的基因沉默。
siPOOLs产品优势
- 使用简单快捷:siPOOLs与多种转染试剂兼容,几天内就能看到结果。
- 高度特异性且有效:siPOOLs在标准细胞系中将脱靶率降低5-25 倍,且在1-3 nM下实现基因敲低率≥70%。
- 一致的表型:由序列非依赖性的siPOOL产生的表型高度一致。
- 确保经过检验:通过RT-qPCR进行siPOOL验证,如果在最佳转染下敲低率低于70%,则有可能重新设计。
- 使用专业数据库的注释进行定制设计:专业的设计,确保优化热力学特性且避免旁系同源基因。
- HPLC纯化且无毒:所有siPOOL均经过HPLC纯化,可降低污染物和副作用的风险。
关键问题:siRNAs的脱靶效应
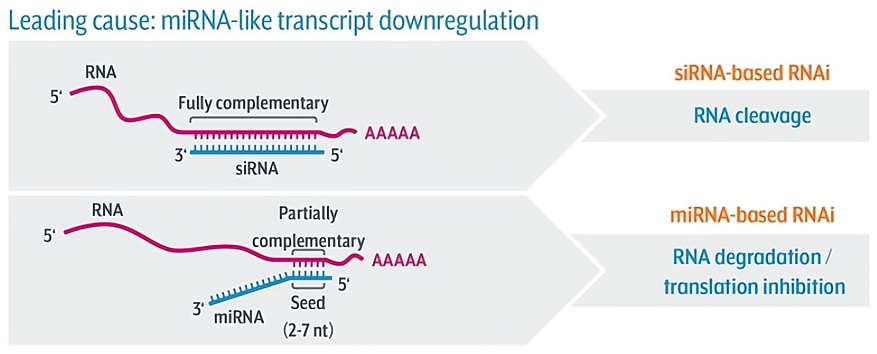
siRNA通常与靶RNA转录本互补结合,通过RNAi机制指导其降解。脱靶效应主要是由siRNA模拟内源性基因调节因子microRNA(miRNA)引起的。由于miRNA只需要6个碱基种子匹配到3'非翻译区(UTR),即可触发转录本下调。因此,siRNA在通过这种机制发挥作用时,可能会改变许多意想不到的靶标基因的表达。
siPOOLs如何提高特异性?
单个siRNA或含有3-4个siRNA的低复杂度siRNA库,常常击中多个脱靶基因,并表现出易变的靶基因敲低效率。 siPOOL是高度复杂且确定的30个siRNA池,每个siRNA以皮摩尔工作浓度存在。因此,siPOOLs :
1. 有效稀释了每个siRNA的脱靶特征,提高了靶向特异性。
2. 确保了靶基因的协同敲低,产生更稳健、更可靠的结果。
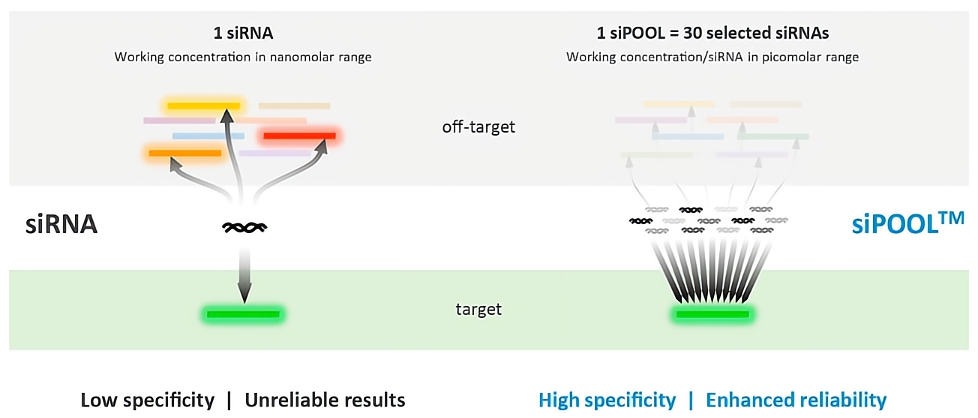
使用siPOOL具有更高的特异性
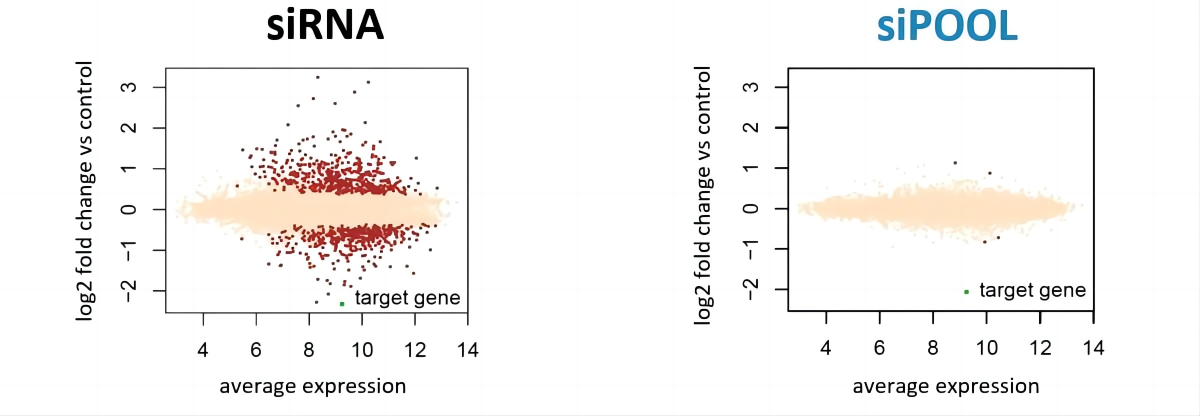
特异性好的试剂,应仅影响其靶标基因。
HeLa细胞的微阵列表达谱分析显示,单个siRNA可以诱导许多脱靶基因(红点),而针对同一靶基因(绿点)的siPOOL,或许也包含有非特异性的siRNA,但是大大降低了脱靶效应。
通过siPOOL实现更好的重现性和有效的敲低效率
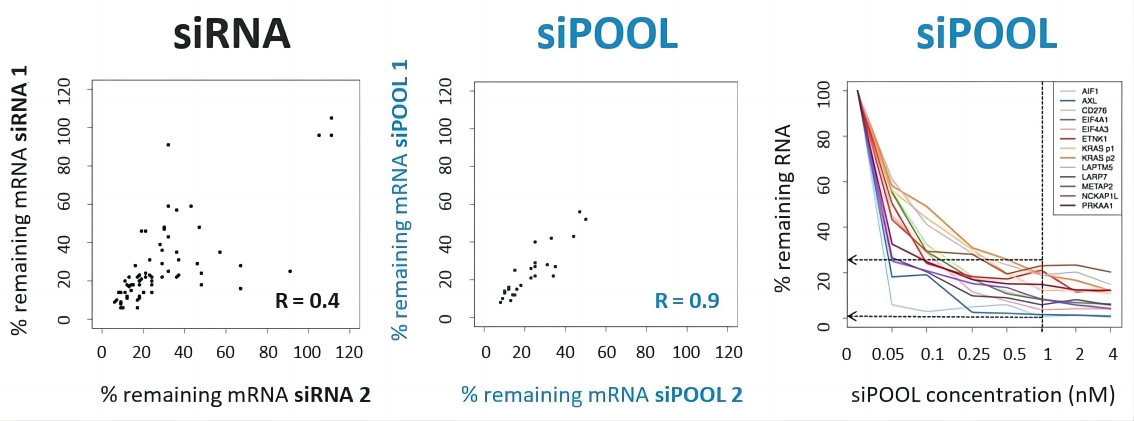
普通siRNA的敲低效率差异很大。
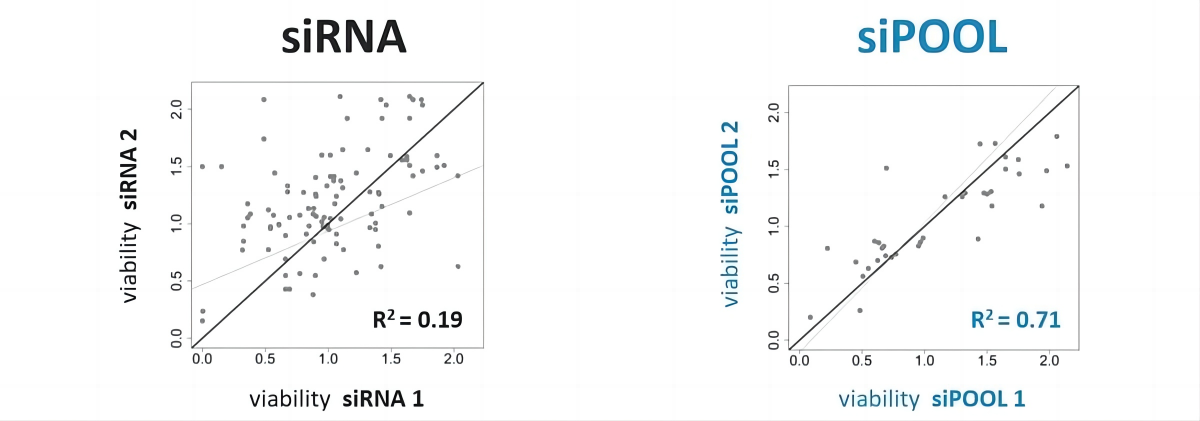
与单个siRNA相比,针对同一基因的siPOOLs具有相似的敲低效率,表明具有更高的稳健性和可重复性(左图和中图:靶RNA水平的实时定量PCR测量的相关图)。 siPOOLs也表现出有效的基因敲低。在常用的细胞系中,许多基因通常在1 nM浓度下实现75-98%的基因敲低效率(右图)。
您可以信赖的表型
如果RNAi效果可靠且特异,靶向同一基因的两种试剂应产生相似的表型。
每个基因分别使用两个siPOOL池,一个来自我们的人类激酶siPOOL文库,一个是市售的每个基因含3条siRNAs的文库,科学家在A549细胞中筛选了36个基因并检测了细胞活力,结果表明siPOOLs产生了较好的一致性表型。
siTOOLs Biotech由Michael Hannus博士和Gunter Meister教授于2013年9月成立,由德国雷根斯堡大学 和Intana Bioscience制药服务公司联合运营。目前推出有3个产品系列:(1)siPOOLs:一种siRNA混合物,有效增加基因沉默特异性,“稀释”传统RNA干扰试剂的脱靶效应。(2)raPOOLs:一种稳健的RNA亲和纯化方案,适用于基因功能和相互作用的生物化学研究。(3)riboPOOLs:适合任意物种的核糖体RNA去除试剂,高效且经济。
siTOOLs Biotech始终致力于帮助学术研究人员、科学家、制药/生物技术公司和RNAi筛选人员。目前,已帮助许多论文发表在《自然》、《细胞》、《自然医学》、《科学报告》等顶级学术期刊上。期待与您的合作,我们将为您的RNA研究提供量身定制的分子工具。
Hannus M. et al. (2014) siPools: highly complex butaccurately defined siRNA pools eliminate off-targeteffects. Nucleic Acids Res 42(12): 8049-61
Zimmermann et al. (2023) Cold Atmospheric Plasma TriggersApoptosis via the Unfolded Protein Response in MelanomaCells MDPI
Müller et al. (2023) Plakophilin 3 facilitates G1/S phasetransition and enhances proliferation by capturing RB protein inthe cytoplasm and promoting EGFR signaling Cell Reports
Luo et al. (2023) The long non-coding RNA LINC00958 isinduced in psoriasis epidermis and modulates epidermalproliferation Journal of Investigative Dermatology
Staebler et al. (2023) MIA/CD-RAP Regulates MMP13 and Is aPotential New Disease-Modifying Target for OsteoarthritisTherapy MDPI
Hertel et al. (2022) USP32-regulated LAMTOR1ubi impacts mTORC1 activation and autophagyinduction Cell Reports
Yavon et al. (2022) High-throughput morphometric andtranscriptomic profiling uncovers composition of naïve andsensory-deprived cortical cholinergic VIP/CHAT neurons TheEMBO Journal
Kim et al. (2022) Screening of the siGPCR library incombination with cisplatin against lung cancers ScientificReports
Mitani et al. (2022) SNAP23-Mediated Perturbation ofCholesterol-Enriched Membrane Microdomain Promotes ExtracellularVesicle Production in Src-Activated Cancer Cells Biologicaland Pharmaceutical Bulletin
Blümel et al. (2022) Primary cilia contribute to theaggressiveness of atypical teratoid/rhabdoid tumors Cell Death& Disease
Rovira et al. (2022) The lysosomal proteome of senescentcells contributes to the senescence secretome Aging Cell
Koch et al. (2022) Targeting the Retinoblastoma/E2Frepressive complex by CDK4/6 inhibitors amplifies oncolytic potencyof an oncolytic adenovirus Nature Communications
Schaefer et al. (2022) Global and precise identificationof functionalmiRNA targets in mESCs by integrativeanalysis EMBO Reports
Kerstin Dörner et al. (2022) Genome-wide RNAi screenidentifies novel players in human 60S subunit biogenesis includingkey enzymes of polyamine metabolism Nucleic Acids Research
Bartl et al. (2022) The HHIP-AS1 lncRNA promotestumorigenicity through stabilization of dynein complex 1 in humanSHH-driven tumors Nature Communications
Banas et al. (2022) The Interplay of NEAT1 and miR-339-5pInfluences on Mesangial Gene Expression and Function in VariousDiabetic-Associated Injury Models non-coding RNA
Mestre-Farràs et al. (2022) Melanoma RBPome identificationreveals PDIA6 as an unconventional RNA-binding protein involved inmetastasis Nucleic Acids Research
Dessauges et al. (2022) Optogenetic actuator – ERKbiosensor circuits identify MAPK network nodes that shape ERKdynamics Molecular Systems Biology
Falke et al. (2022) Knockdown of the stem cell markerMusashi-1 inhibits endometrial cancer growth and sensitizes cellsto radiation Stem Cell Research & Therapy
Gutierrez-Prat et al. (2022) DUSP4 protects BRAF- andNRAS-mutant melanoma from oncogene overdose through modulation ofMITF Life Science Alliance
Lechner et al. (2022) Target deconvolution of HDACpharmacopoeia reveals MBLAC2 as common off-target NatureChemical Biology
Daniel Kummer et al. (2022) A JAM-A–tetraspanin–αvβ5integrin complex regulates contact inhibition oflocomotion Journal of Cell Biology
Delicato, A. et al (2022) YB-1 Oncoprotein ControlsPI3K/Akt Pathway by Reducing Pten Protein Level Genes
Szachnowski et al. (2022) Transcriptomic landscapes ofSARS-CoV-2-infected and bystander lung cells reveal a selectiveupregulation of NF-κB-dependent coding and non-coding proviraltranscripts bioRxiv - preprint
Hauffe, L. et al. (2022) Eukaryotic translation initiationfactor 4E binding protein 1 (EIF4EBP1) expression in glioblastomais driven by ETS1- and MYBL2-dependent transcriptionalactivation Cell Death Discovery
Till Braun et al. (2022) Non-canonical function ofAGO2 augments T-cell receptor signaling in T-cell prolymphocyticleukemia Amercian Association for Cancer Research
Karabid et al. (2022) Angpt2/Tie2 autostimulatoryloop controls tumorigenesis EMBO Mol Med
X. Li et al. (2022) 5’isomiR-183-5p|+2 Elicits TumorSuppressor Activity in a Negative Feedback Loop withE2F1 Journal of Experimental & Clinical Cancer Research -preprint -
Thölmann et al. (2022) JAM-A interacts with α3β1 integrinand tetraspanins CD151 and CD9 to regulate collective cellmigration of polarized epithelial cells Cellular and MolecularLife Sciences
Thea Reinkens et al. (2021) Ago-RIP Se IdentifiesNew MicroRNA-449a-5p Target Genes Increasing SorafenibEfficacy in Hepatocellular Carcinoma Journal OfCancer
Weihua Qin et. Al (2022) Probing protein ubi inlive cells bioRxiv - preprint
Belitškin et al. (2021) Hepsin regulates TGFβ signalingvia fibronectin proteolysis EMBO reports
Q. Zou et al (2021) Utility of single-shot compressedsensing cardiac magnetic resonance cine imaging for assessment ofbiventricular function in free-breathing and arrhythmic pediatricpatients International Journal of Cardiology
J. Laisney et al (2021) Delivery of short hairpin RNA inthe neotropical brown stink bug, Euschistus heros, using acomposite nanomaterial Pesticide Biochemistry andPhysiology
Indacochea et al (2021) Cold-inducible RNA bindi ngprotein promotes breast cancer cell maligna ncy by regulating Cystatin C levels Cold Spring Harbor Laboratory Press
Khan et al (2021) Developing Tumor RadiosensitivitySignatures Using LncRNAs Radiation Research
Ghodke et al (2021) AHNAK controls 53BP1-mediated p53response by restraining 53BP1 oligomerization and phaseseparation Molecular Cell
Gaza et al (2021) Identification of novel targets ofmiR-622 in hepatocellular carcinoma reveals common regulation ofcooperating genes and outlines the oncogenic role of zinc fingerCCHC-type containing 11 Neoplasia
Indacochea et al (2021) Cold-inducible RNA binding proteinpromotes breast cancer cell maligna ncy by regulating Cys tatin Clevels Cold Spring Harbor Laboratory Press
Mercier et al (2020) Endosomal membrane tension regulatesESCRT-III-dependent intra-lumenal vesicle formation NatureCell Biology
Ghosh et al (2020) Prevention of dsRNA-induced interferonsignaling by AGO1x is linked to breast cancer cellproliferation The EMBO Journal
Dhamija et. al (2020) A pan-cancer analysis revealsnonstop extension mutations causing SMAD4 tumour suppressordegradation Nature Cell Biology
Gandhi et. al (2020) The lncRNA lincNMR regulatesnucleotide metabolism via a YBX1 - RRM2 axis in cancer NatureCommunications
Stephan et. al (2020) MICOS assembly controlsmitochondrial inner membrane remodeling and crista junctionredistribution to mediate cristae formation The EMBOjournal
Liebig et. al (2020) HuRdling Senescence: HuR BreaksBRAF-Induced Senescence in Melanocytes and Supports MelanomaGrowth. MDPI Cancers
Prasad et. al (2020) The UPR sensor IRE1α and theadenovirus E3-19K glycoprotein sustain persistent and lyticinfections. Nature Communications
Marchetto et. al (2020) Oncogenic hijacking of adevelopmental transcription factor evokes vulnerability towardoxidative stress in Ewing sarcoma. NatureCommunications
Larios, J. et. al (2020) ALIX- and ESCRT-III–dependentsorting of tetraspanins to exosomes. JCB
Mahli, A. et. al (2019) Bone Morphogenic Protein-8BExpression is Induced in Steatotic Hepatocytes and Promotes HepaticSteatosis and Inflammation in Vitro. Cells
Renner, K. et. al (2019) Restricting Glycolysis PreservesT Cell Effector Functions and Augments CheckpointTherapy. Cell Rep
Werner, S. et. al (2019) MRTF-A controls myofibroblasticdifferentiation of human multipotent stromal cells and theirtumour-supporting function in xenograft models. ScientificReports
Braun, C. et. al (2019) Inhibition of peptidyl-prolylisomerase (PIN1) and BRAF signaling to target melanoma. Am JTransl Res
Kappelmann-Fenzl, M. et. al (2019) C-Jun drives melanomaprogression in PTEN wild type melanoma cells. Cell DeathDis
Simchovitz, A et. al (2019) NEAT1 is overexpressed inParkinson’s disease substantia nigra and confers drug-inducibleneuroprotection from oxidative stress. FASEB J
Mahli, A et. al (2019) Bone Morphogenetic Protein-8BExpression is Induced in Steatotic Hepatocytes and Promotes HepaticSteatosis and Inflammation In Vitro. Cells
Dietrich, P et. al (2019) The Delta Subunit ofRod-Specific Photoreceptor cGMP Phosphodiesterase (PDE6D)Contributes to Hepatocellular CarcinomaProgression. Cancers
Feuerer, L et. al (2019) Role of MIA (melanoma inhibitoryactivity) in melanocyte senescence. Pigment Cell MelanomaRes
Ziegler, C. et. al (2019) The long non‐coding RNALINC00941 and SPRR5 are novel regulators of human epidermalhomeostasis. EMBO Rep
Choi, K. et. al (2019) Binary Targeting of siRNA toHematologic Cancer Cells In Vivo Using Layer‐by‐LayerNanoparticles. Adv Funct Mater
Spies, J. et. al (2019) 53BP1 nuclear bodies enforcereplication timing at under-replicated DNA to limit heritable DNAdamage. Nat Cell Biol
Dietrich, P. et al. (2019) Neuroblastoma RAS ViralOncogene Homolog (NRAS) Is a Novel Prognostic Marker andContributes to Sorafenib Resistance in HepatocellularCarcinoma. Neoplasia
Vendramin, R. et al. (2018) SAMMSON fosters cancer cellfitness by concertedly enhancing mitochondrial and cytosolictranslation. Nat Struct Mol Biol
Coscia, F. et al. (2018) Multi-level Proteomics IdentifiesCT45 as a Chemosensitivity Mediator and Immunotherapy Target inOvarian Cancer. Cell
Kästle, M. et al. FKBP51 modulates steroid sensitivity andNFκB signalling: A novel anti-inflammatory drug target. Eur JImmunol
Stieglitz, D. et al. BMP6-induced modulation of the tumormicro-milieu. Oncogene 10.1038/s4-x(2018)
Weber, A., Schwarz, S. C. et al. Epigenome-wide DNAmethylation profiling in Progressive Supranuclear Palsy revealsmajor changes at DLX1. Nat Commun 9, 2929 (2018)
Noethel, B. et al. Transition of responsivemechanosensitive elements from focal adhesions to adherensjunctions upon epithelial differentiation. Mol BiolCell 10.1091/mbc.E17-06-0387(2018)
Prestigiacomo, V., and Suter-Dick, L. Nrf2 protectsstellate cells from Smad-dependent cell activation. PLoSOne 13, e0201044 (2018)
Vallin, B. et al. Novel short isoforms of adenylyl cyclaseas negative regulators of cAMP production. Biochim BiophysActa - Mol Cell Res (2018)
Rietscher, K. et al.14-3-3 Proteins Regulate DesmosomalAdhesion via Plakophilins.” Journal of cellscience 131(10). (2018).
Jiang, P. et al. Genome-Scale Signatures of GeneInteraction from Compound Screens Predict Clinical Efficacy ofTargeted Cancer Therapies.” Cell Systems 6(3):343–354.e5. (2018).
So, D. et al. Cervical Cancer Is Addicted to SIRT1Disarming the AIM2 AntiviralDefense. Oncogene (2018).
Klingenberg, M. et al. The lncRNA CASC9 and RNA BindingProtein HNRNPL Form a Complex and Co-Regulate Genes Linked to AKTSignaling. Hepatology (2018).
Polycarpou-Schwarz, M. et al. The Cancer-AssociatedMicroprotein CASIMO1 Controls Cell Proliferation and Interacts withS Epoxidase Modulating Lipid DropletFormation. Oncogene (2018)
Zeiner, P. S. et al. CD74 regulates complexity of tumorcell HLA class II peptidome in brain metastasis and is a positiveprognostic marker for patient survival. Acta Neuropathol.Commun. 6, 18 (2018).
Ziegelmann, B et al. Lithium chloride effectively killsthe honey bee parasite Varroa destructor by a systemic mode ofaction. Scientific Reports 8(1), 683 (2018)
Dietrich, P. et al. Wild type Kirsten rat sarcoma is anovel microRNA-622-regulated therapeutic target for hepatocellularcarcinoma and contributes to sorafenibresistance. Gut (2017)
Dietrich, P. et al. Wild-type KRAS is a novel therapeutictarget for melanoma contributing to primary and acquired resistanceto BRAF inhibition. Oncogene (2017)
Kaller, M. et al. Loss of p53-inducible long non-codingRNA LINC01021 increases chemosensitivity. Oncotarget 8,102783–102800 (2017).
Ilina, E. I. et al. Effects of soluble CPE on glioma cellmigration are associated with mTOR activation and enhanced glucoseflux. Oncotarget 8, 67567–67591 (2017).
Goyal, A. et al. A cautionary tale of sense-antisense genepairs: independent regulation despite inverse correlation ofexpression. Nucleic Acids Research (2017)http://dx.doi.org/10.1093/nar/gkx952
Steinbach, A. et al. ERAP1 overexpression in HPV-inducedmalignancies: A possible novel immune evasionmechanism. Oncoimmunology 6, e1336594 (2017).
Sluch, V. M. et al. Enhanced Stem Cell Differentiation andImmunopurification of Genome Engineered Human Retinal GanglionCells. STEM CELLS Translational Medicine. (2017)doi:10.1002/sctm.17-0059
Armento, A. et al. Carboxypeptidase E transmits itsanti-migratory function in glioma cells via transcriptionalregulation of cell architecture and motility regulatingfactors. International Journal of Oncology 51(2), 702-714(2017)
Nötzold, L. et al. The long non-coding RNA LINC00152 isessential for cell cycle progression through mitosis in HeLacells. Scientific Reports 7, 2265 (2017) Also citesraPOOL
Welsbie, D. S. et al. Enhanced Functional GenomicScreening Identifies Novel Mediators of Dual Leucine ZipperKinase-Dependent Injury Signaling inNeurons. Neuron 94(6), 1142–1154.e6 (2017)
Ruivo, M. T. G. et al. Host AMPK Is a Modulator ofPlasmodium Liver Infection. Cell Rep. 1–7 (2017).
Treiber, T. et al. A Compendium of RNA-Binding Proteinsthat Regulate MicroRNA Biogenesis. Mol. Cell 66,270–284.e13 (2017).
Seiler, J. et al. The lncRNA VELUCT strongly regulatesviability of lung cancer cells despite its extremely lowabundance. Nucleic Acids Res. (2017)
Goyal, A. et al. Challenges of CRISPR/Cas9 applicationsfor long non-coding RNA genes. Nucleic AcidsRes. (2016)
Ott, C. A. et al. Induction of exportin-5 expressionduring melanoma development supports the cellular behavior of humanmalignant melanoma cells. Oncotarget 7, 62292–62304(2016).
Adriaens, C. et al. p53 induces formation of NEAT1lncRNA-containing paraspeckles that modulate replication stressresponse and chemosensitivity. Nat. Med. (2016).
Coll-Bonfill, N. et al. Slug is increased in vascularremodeling and induces a smooth muscle cell proliferativephenotype. PLoS One 11, 1–21 (2016).
Kordaß, T. et al. SOX5 is involved in balanced MITFregulation in human melanoma cells. BMC Med. Genomics 9,10 (2016).
Hauptmann, J. et al. Biochemical isolation of Argonauteprotein complexes by Ago-APP. Proc. Natl. Acad. Sci. 112,11841–11845 (2015).
Blirando, K. et al. The stellate vascular smooth musclecell phenotype is induced by IL-1β via the secretion of PGE2 andsubsequent cAMP-dependent protein kinase Aactivation. Biochim. Biophys. Acta. (2015).
Tuncay, H. et al. JAM-A regulates cortical dyneinlocalization through Cdc42 to control planar spindle orientationduring mitosis. Nat. Commun. 6, 8128 (2015).
Schraivogel, D. et al. Importin-β facilitates nuclearimport of human GW proteins and balances cytoplasmic gene silencingprotein levels. Nucleic Acids Res. (2015)
Schönemann L. et al.: Reconstitution of CPSF active inpolyadenylation: recognition of the polyadenylation signal byWDR33. Genes Dev. 28(21), 2381-93 (2014)
Prasad, V. et al. Chemical induction of unfolded proteinresponse enhances cancer cell killing through lytic virusinfection. J. Virol. 88, 13086–13098 (2014).
Patrick C.H. Lo, PhD: siRNAs: Jumping in the Pool to AvoidHitting Innocent Bystanders. BioTechniques (2014)
Thalyana S. Advances in RNAi Tools andTechnologies. Genetic Engineering & BiotechnologyNews. April 33(8): 20-22, 24 (2013)




















 1356
1356











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








