数据集:cellDancer文件 PancreaticEndocrinogenesis_cellDancer_estimation.csv
import random
import numpy as np
import pandas as pd
import numpy as np
import scipy
import warnings
warnings.filterwarnings('ignore')
import dynamo as dyn
from celldancer.utilities import export_velocity_to_dynamo1.将cellDancer的预测输入dynamo
# First create dynamo adata
adata_dyn = dyn.sample_data.pancreatic_endocrinogenesis()
pancreas_genes = ["Hes1","Nkx6-1","Nkx2-2","Neurog3","Neurod1","Pax4","Pax6",
"Arx","Pdx1","Ins1","Ins2","Ghrl","Ptf1a","Iapp","Isl1", "Sox9","Gcg"]
dyn.pp.recipe_monocle(adata_dyn, n_top_genes=4000,
fg_kwargs={"shared_count": 20},
genes_to_append=pancreas_genes)
dyn.tl.dynamics(adata_dyn, model='static')
# replace adata.layers['velocity_S'], adata.var['use_for_dynamics'], and adata.var['use_for_transition'] with cellDancer outputs.
cellDancer_df = pd.read_csv('your_path/PancreaticEndocrinogenesis_cellDancer_estimation.csv')
adata = export_velocity_to_dynamo(cellDancer_df,adata_dyn)2.将 RNA 速率投影到嵌入空间上
pancreas_cluster_cmap = {'Ductal': '#3361A5','Ngn3 low EP': '#248AF3',
'Ngn3 high EP': '#14B3FF',
'Pre-endocrine': '#88CEEF','Alpha': '#ff4800',
'Beta': '#B81136','Delta': 'green',
'Epsilon': '#03B3B0'}
dyn.tl.reduceDimension(adata, n_pca_components=30)
dyn.tl.cell_velocities(adata, method="pearson",
other_kernels_dict={"transform": "sqrt"})
dyn.tl.cell_velocities(adata, basis="pca")
dyn.pl.streamline_plot(adata, color=["clusters"], basis="umap",
color_key = pancreas_cluster_cmap,
show_legend="on data",
show_arrowed_spines=True)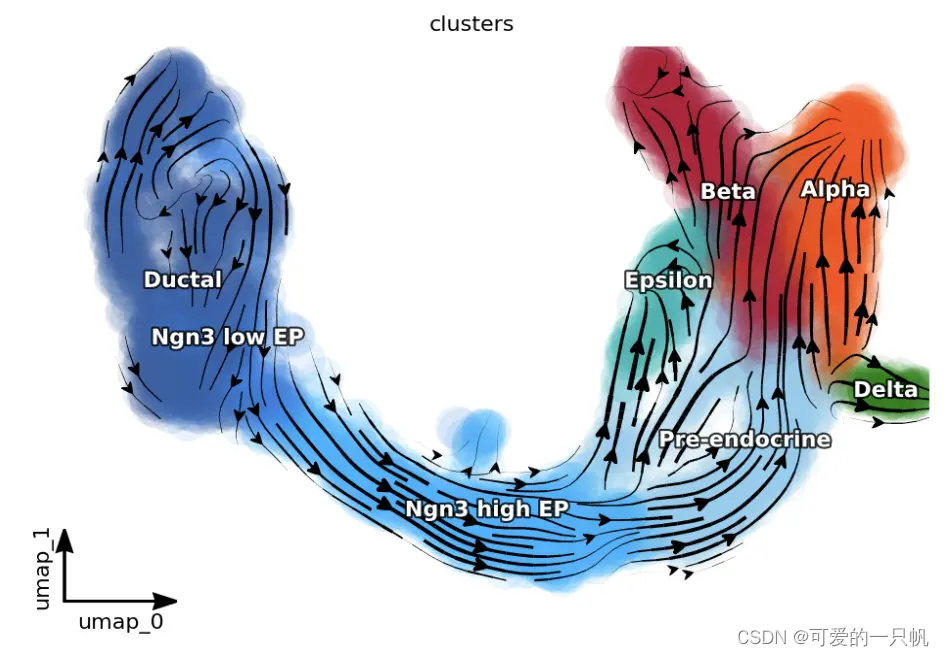
3.学习和可视化 UMAP 上的矢量场
dyn.vf.VectorField(adata, basis="pca", pot_curl_div=True)
dyn.vf.VectorField(adata, basis="umap", pot_curl_div=True)
dyn.vf.speed(adata, basis="pca")
dyn.vf.divergence(adata, basis="pca")
dyn.vf.acceleration(adata, basis="pca")
dyn.vf.curl(adata, basis="umap")
dyn.pl.topography(adata, color=['clusters'], basis='umap',
background='white',
streamline_color='black',
color_key = pancreas_cluster_cmap,
show_legend='on data', terms=("streamline"))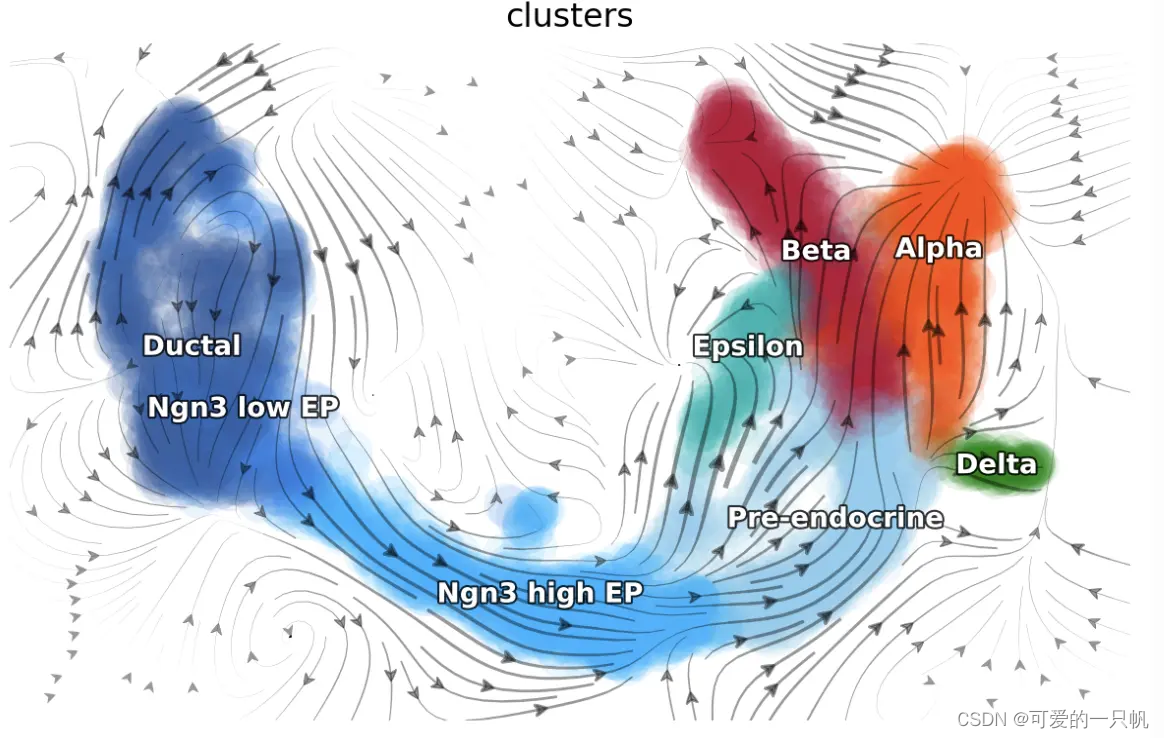
adata_load_fix_points=adata.copy()
Xss, ftype, conf = adata_load_fix_points.uns['VecFld_umap']['Xss'],\
adata_load_fix_points.uns['VecFld_umap']['ftype'],\
adata_load_fix_points.uns['VecFld_umap']['confidence']
fixed_points = [10,7,16,12]
adata_load_fix_points.uns['VecFld_umap']['Xss'] = Xss[fixed_points]
adata_load_fix_points.uns['VecFld_umap']['ftype'] = ftype[fixed_points]
adata_load_fix_points.uns['VecFld_umap']['confidence']=conf[fixed_points]
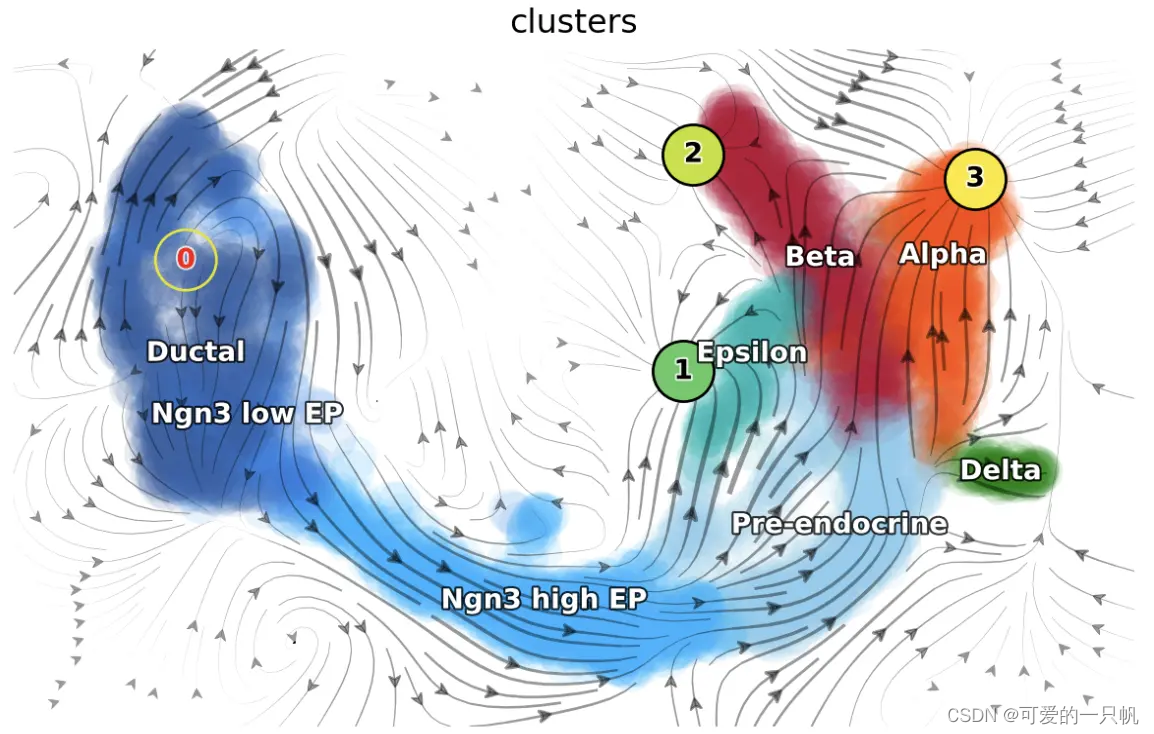
dyn.pl.topography(
adata_load_fix_points,
markersize=500,
basis="umap",
fps_basis="umap",
color_key = pancreas_cluster_cmap,
color=["clusters"],
show_legend='on data',
save_show_or_return='show'
)
4.Jacobian分析检测基因调控
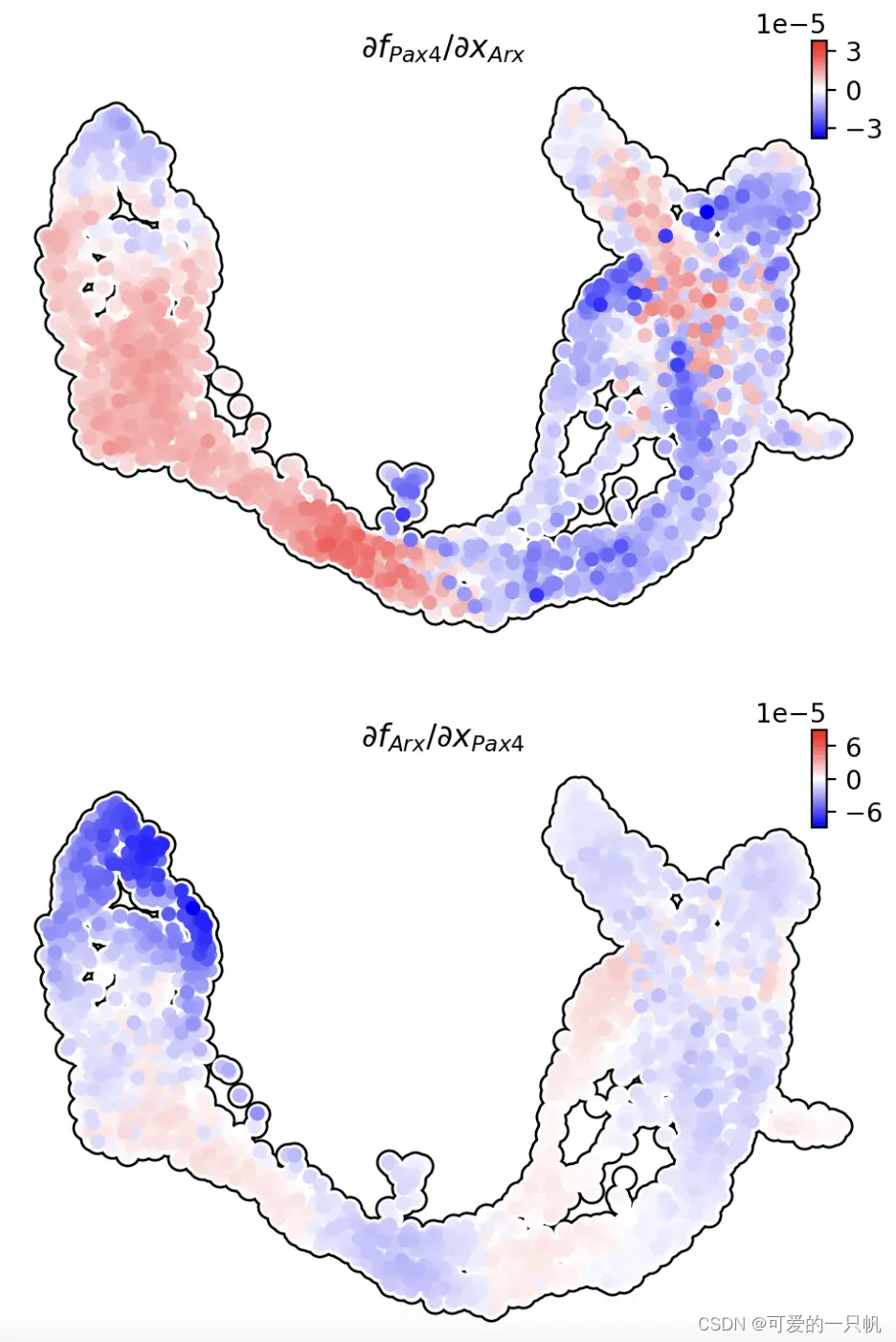
dyn.vf.jacobian(adata, regulators=['Arx','Pax4'])
dyn.pl.jacobian(
adata,
basis="umap",
regulators=['Arx'],
effectors=['Pax4'],
alpha=1,
)
dyn.pl.jacobian(
adata,
basis="umap",
regulators=['Pax4'],
effectors=['Arx'],
alpha=1,
)
在UMAP上绘制特定基因的表达
dyn.pl.umap(adata, color=["Arx",'Pax4'],save_show_or_return='show') 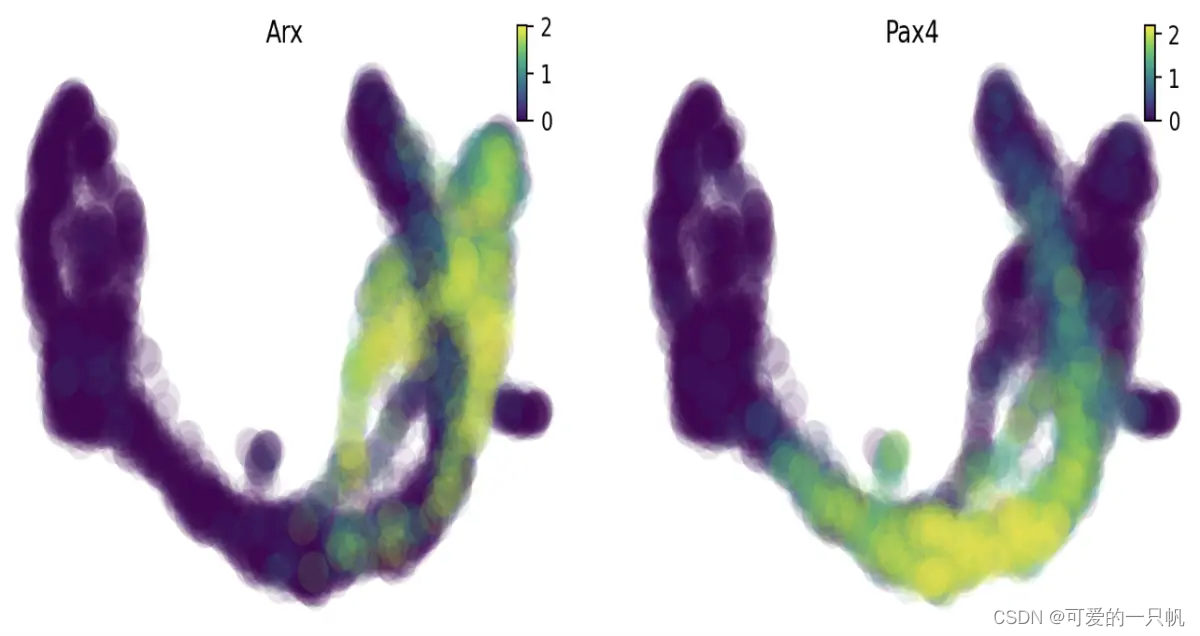
参考:Downstream analysis using Dynamo — cellDancer documentation (guangyuwanglab2021.github.io)
Cell, 2022, 185(4):690-711.e45.







 本文介绍了如何将cellDancer的预测结果导入Dynamo,包括处理数据、在UMAP上投影RNA速率、学习和可视化矢量场、进行Jacobian分析以及基因表达可视化。这些步骤展示了如何利用Dynamo进行下游数据分析,以理解胰腺细胞发育过程中的动态变化。
本文介绍了如何将cellDancer的预测结果导入Dynamo,包括处理数据、在UMAP上投影RNA速率、学习和可视化矢量场、进行Jacobian分析以及基因表达可视化。这些步骤展示了如何利用Dynamo进行下游数据分析,以理解胰腺细胞发育过程中的动态变化。














 1816
1816











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








