
简 介
斜随机生存森林(ORSF)是一种集成方法,用于右删节存活数据,它使用输入变量的线性组合递归地划分一组训练数据。正则化Cox比例风险模型用于识别每个递归划分步骤中输入变量的线性组合。模拟和真实数据的基准测试结果表明,与随机生存森林、条件推理森林、回归和增强相比,ORSF预测的风险函数具有较高的预测价值。在Jackson心脏研究数据的应用中,使用ORSF证明了变量和部分依赖性,并强调了其10年预测动脉粥样硬化性心血管疾病事件(ASCVD;中风,冠心病)。根据ORSF、条件推理森林和合并队列风险方程,展示了比较变量和部分效应估计的可视化效果。

软件包安装
软件包安装两种方式:
install.packages("devtools")
install.packages("remotes")
remotes::install_github("ropensci/aorsf")
install.packages('aorsf')数据读取
数据集来自survival包中的pbc,来自梅奥诊所在1974年至1984年间进行的PBC试验。在这10年期间,共有424名PBC患者转至梅奥诊所,符合D-青霉胺随机安慰剂对照试验的资格标准。数据集中的前312例患者参加了随机试验,数据基本完整。另外112例患者没有参加临床试验,但同意记录基本测量值并随访以维持生存。其中6例在诊断后不久就失去了随访,所以这里的数据是额外的106例病例以及312名随机参与者。
age: in years
albumin: serum albumin (g/dl)
alk.phos: alkaline phosphotase (U/liter)
ascites: presence of ascites
ast: aspartate aminotransferase, once called SGOT (U/ml)
bili: serum bilirunbin (mg/dl)
chol: serum cholesterol (mg/dl)
copper: urine copper (ug/day)
edema: 0 no edema, 0.5 untreated or successfully treated 1 edema despite diuretic therapy
hepato: presence of hepatomegaly or enlarged liver
id: case number
platelet: platelet count
protime: standardised blood clotting time
sex: m/f
spiders: blood vessel malformations in the skin
stage: histologic stage of disease (needs biopsy)
status: status at endpoint, 0/1/2 for censored, transplant, dead
time: number of days between registration and the earlier of death,transplantion, or study analysis in July, 1986
trt: 1/2/NA for D-penicillmain, placebo, not randomised
trig: triglycerides (mg/dl)
library(aorsf)
library(tidyverse)
pbc <- pbc_orsf
# An oblique survival RF
head(pbc)
## id time status trt age sex ascites hepato spiders edema bili
## 1 1 400 1 d_penicill_main 58.76523 f 1 1 1 1 14.5
## 2 2 4500 0 d_penicill_main 56.44627 f 0 1 1 0 1.1
## 3 3 1012 1 d_penicill_main 70.07255 m 0 0 0 0.5 1.4
## 4 4 1925 1 d_penicill_main 54.74059 f 0 1 1 0.5 1.8
## 5 5 1504 0 placebo 38.10541 f 0 1 1 0 3.4
## 7 7 1832 0 placebo 55.53457 f 0 1 0 0 1.0
## chol albumin copper alk.phos ast trig platelet protime stage
## 1 261 2.60 156 1718.0 137.95 172 190 12.2 4
## 2 302 4.14 54 7394.8 113.52 88 221 10.6 3
## 3 176 3.48 210 516.0 96.10 55 151 12.0 4
## 4 244 2.54 64 6121.8 60.63 92 183 10.3 4
## 5 279 3.53 143 671.0 113.15 72 136 10.9 3
## 7 322 4.09 52 824.0 60.45 213 204 9.7 3pbc = na.omit(pbc)
table(pbc$status)
##
## 0 1
## 165 111library(sampling)
set.seed(123)
# 每层抽取70%的数据
train_id <- strata(pbc, "status", size = rev(round(table(pbc$status) * 0.7)))$ID_unit
# 训练数据
trainData <- pbc[train_id, ]
# 测试数据
testData <- pbc[-train_id, ]实例操作
构建模型
pbc_fit <- orsf(data = trainData, n_tree = 20, formula = Surv(time, status) ~ . -
id)
res <- orsf_summarize_uni(pbc_fit)
library(data.table)
head(as.data.table(res))
## variable importance value mean medn lwr upr
## <char> <num> <char> <num> <num> <num> <num>
## 1: bili 0.5172414 0.60 0.2663037 0.1932877 0.05741174 0.4269579
## 2: bili 0.5172414 0.80 0.2724312 0.1996642 0.06232726 0.4480355
## 3: bili 0.5172414 1.60 0.3080398 0.2189249 0.08703489 0.5017554
## 4: bili 0.5172414 3.70 0.4404167 0.3980655 0.20825906 0.6614583
## 5: bili 0.5172414 7.18 0.5075947 0.5301124 0.28791330 0.7291667
## 6: copper 0.4909091 27.2 0.2997513 0.2107370 0.06277056 0.5096297
## pred_horizon level
## <num> <char>
## 1: 1741 <NA>
## 2: 1741 <NA>
## 3: 1741 <NA>
## 4: 1741 <NA>
## 5: 1741 <NA>
## 6: 1741 <NA>验证
预测predict()参数中pred_type有7种选择:none, surv, risk, chf, mort, leaf, or time, 可以获得预测生存矩阵和相对风险:
# predicted risk, the default
predict(pbc_fit, new_data = testData, pred_type = "risk", pred_horizon = c(360, 720,
1800))
# predicted survival, i.e., 1 - risk
predict(pbc_fit, new_data = testData, pred_type = "surv", pred_horizon = c(360, 720,
1800))
# predicted cumulative hazard function (expected number of events for person i
# at time j)
predict(pbc_fit, new_data = testData, pred_type = "chf", pred_horizon = c(360, 720,
1800))
# Predict mortality
predict(pbc_fit, new_data = testData, pred_type = "mort")变量重要性
针对变量分析包括 :
变量的重要性(Variable Importance);
变量的选择(Variable selection);
变量的互作(Variable Interactions)。
# Variable Importance
orsf_vi(pbc_fit)
## bili copper ascites edema age protime albumin
## 0.51724138 0.49090909 0.47222222 0.36834823 0.34545455 0.32075472 0.24444444
## ast platelet hepato trig chol stage spiders
## 0.17741935 0.15873016 0.14583333 0.14285714 0.14285714 0.13235294 0.09803922
## sex alk.phos trt
## 0.06818182 0.05263158 0.00000000# Variable selection
orsf_vs(pbc_fit, n_predictor_min = 5)
## n_predictors stat_value variables_included
## <int> <num> <list>
## 1: 5 0.8560700 age,ascites,bili,copper,edema,protime
## 2: 6 0.8484568 age,ascites,bili,chol,copper,edema,...
## 3: 7 0.8381687 age,albumin,ascites,bili,chol,copper,...
## 4: 8 0.8504115 age,albumin,ascites,bili,chol,copper,...
## 5: 9 0.8565844 age,albumin,ascites,bili,chol,copper,...
## 6: 10 0.8525720 age,albumin,ascites,bili,chol,copper,...
## 7: 11 0.8614198 age,albumin,ascites,ast,bili,chol,...
## 8: 12 0.8702675 age,albumin,ascites,ast,bili,chol,...
## 9: 13 0.8534979 age,albumin,alk.phos,ascites,ast,bili,...
## 10: 14 0.8308642 age,albumin,alk.phos,ascites,ast,bili,...
## 11: 15 0.8565844 age,albumin,alk.phos,ascites,ast,bili,...
## 12: 16 0.8412551 age,albumin,alk.phos,ascites,ast,bili,...
## 13: 17 0.8493827 age,albumin,alk.phos,ascites,ast,bili,...
## predictors_included predictor_dropped
## <list> <char>
## 1: age,ascites_1,edema_1,bili,copper,protime protime
## 2: age,ascites_1,edema_1,bili,chol,copper,... chol
## 3: age,ascites_1,edema_1,bili,chol,albumin,... albumin
## 4: age,ascites_1,spiders_1,edema_1,bili,chol,... spiders_1
## 5: age,ascites_1,spiders_1,edema_1,bili,chol,... trig
## 6: age,ascites_1,spiders_1,edema_1,bili,chol,... stage
## 7: age,ascites_1,spiders_1,edema_1,bili,chol,... ast
## 8: age,ascites_1,spiders_1,edema_1,bili,chol,... platelet
## 9: age,ascites_1,spiders_1,edema_1,bili,chol,... alk.phos
## 10: age,ascites_1,hepato_1,spiders_1,edema_1,bili,... hepato_1
## 11: age,ascites_1,hepato_1,spiders_1,edema_0.5,edema_1,... edema_0.5
## 12: age,sex_f,ascites_1,hepato_1,spiders_1,edema_0.5,... sex_f
## 13: trt_placebo,age,sex_f,ascites_1,hepato_1,spiders_1,... trt_placebo# Variable Interactions
interaction <- as.data.frame(orsf_vint(pbc_fit)[, 1:2])
interaction = interaction %>%
separate_wider_delim(interaction, "..", names = c("V1", "V2"))
head(interaction)
## # A tibble: 6 × 3
## V1 V2 score
## <chr> <chr> <dbl>
## 1 ascites edema 2.76
## 2 ascites bili 1.89
## 3 ascites spiders 1.88
## 4 bili stage 1.67
## 5 edema stage 1.53
## 6 edema copper 1.52ggplot(interaction) + geom_point(aes(x = V1, y = V2, color = score, size = score)) +
theme_bw() + labs(y = "", x = "") + theme(axis.text.x = element_text(angle = 90,
vjust = 1, hjust = 1))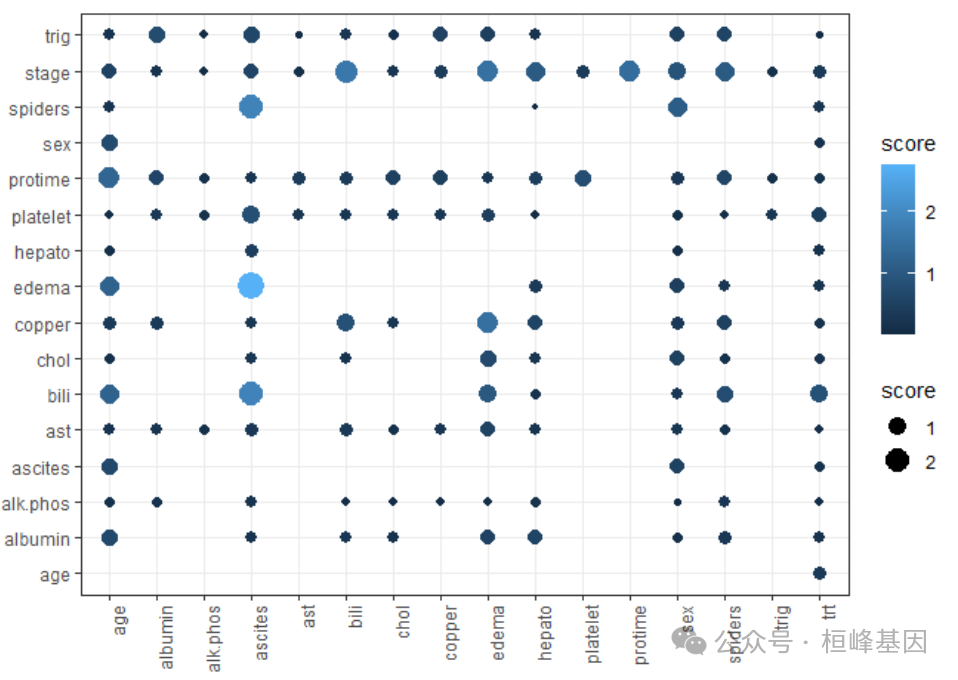
一致性
library(survival)
# 生存分析
pred <- predict(pbc_fit, testData, pred_type = "surv")
str(pred)
## num [1:83, 1] 0.0197 0.3874 0.7159 0.64 0.8972 ...p <- predict(pbc_fit, new_data = testData, pred_type = "risk", pred_horizon = c(360,
720, 1800))
testData$pred = pred
C_index <- data.frame(Cindex = as.numeric(summary(coxph(Surv(time, status) ~ pred,
testData))$concordance[1]))
C_index
## Cindex
## 1 0.7800128生存分析
根据风险值我们可以将患者分为高低风险组,然后绘制生存曲线:
library(survminer)
testData$risk = p[, 1]
group = ifelse(testData$risk > mean(testData$risk), "High", "Low")
f <- survfit(Surv(testData$time, testData$status) ~ group)
f
## Call: survfit(formula = Surv(testData$time, testData$status) ~ group)
##
## n events median 0.95LCL 0.95UCL
## group=High 20 16 1191 611 NA
## group=Low 63 17 3839 3086 NAggsurvplot(f, data = testData, surv.median.line = "hv", legend.title = "Risk Group",
legend.labs = c("Low Risk", "High Risk"), pval = TRUE, ggtheme = theme_bw())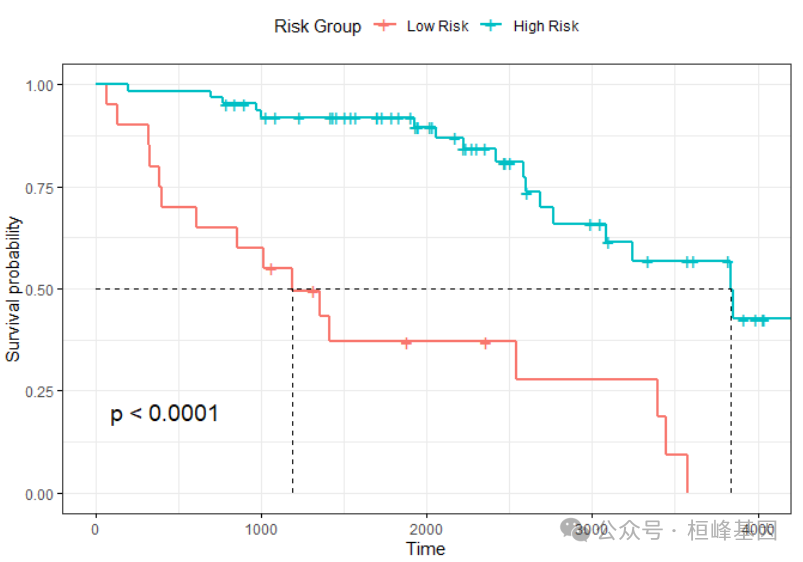
绘制ROC曲线
由于我们所作的模型根时间密切相关因此我们选择timeROC,可以快速的技术出来不同时期的ROC,进一步作图:
library(timeROC)
res <- timeROC(T = testData$time, delta = testData$status, marker = testData$risk,
cause = 1, weighting = "marginal", times = c(360, 720, 1800), ROC = TRUE, iid = TRUE)
res$AUC
## t=360 t=720 t=1800
## 0.7961538 0.8581081 0.8590447confint(res, level = 0.95)$CI_AUC
## 2.5% 97.5%
## t=360 55.39 103.84
## t=720 70.96 100.66
## t=1800 74.68 97.13plot(res, time = 360, col = "red", title = FALSE, lwd = 2)
plot(res, time = 720, add = TRUE, col = "blue", lwd = 2)
plot(res, time = 1800, add = TRUE, col = "green", lwd = 2)
legend("bottomright", c(paste("1 Years ", round(res$AUC[1], 2)), paste("2 Years ",
round(res$AUC[2], 2)), paste("5 Years ", round(res$AUC[3], 2))), col = c("red",
"blue", "green"), lty = 1, lwd = 2)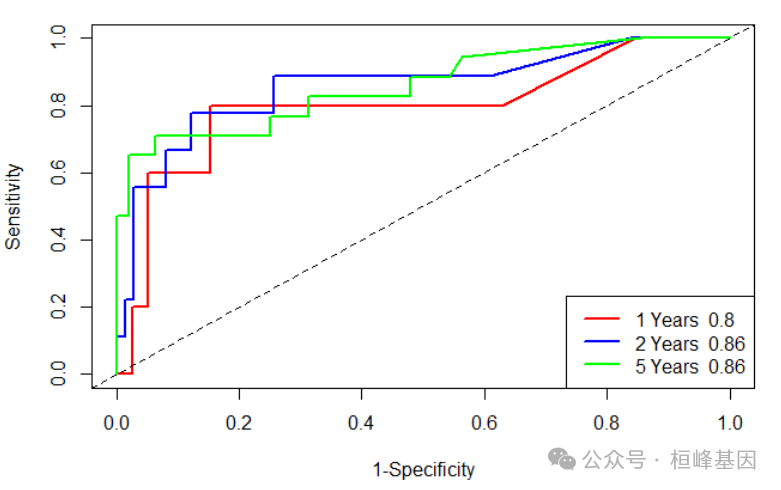
不同时间节点的AUC曲线及其置信区间
再分析不同时间节点的AUC曲线及其置信区间,由于数据量非常小,此图并不明显。
plotAUCcurve(res, conf.int = TRUE, col = "red")
Reference
Jaeger BC, Long DL, Long DM, Sims M, Szychowski JM, Min Y, Mcclure LA, Howard G, Simon N (2019). “Oblique random survival forests.” The Annals of Applied Statistics, 13(3). doi:10.1214/19-aoas1261 https://doi.org/10.1214/19-aoas1261.
Jaeger BC, Welden S, Lenoir K, Speiser JL, Segar MW, Pandey A, Pajewski NM (2023). “Accelerated and interpretable oblique random survival forests.” Journal of Computational and Graphical Statistics, 1-16. doi:10.1080/10618600.2023.2231048 https://doi.org/10.1080/10618600.2023.2231048.
Horst AM, Hill AP, Gorman KB (2020). palmerpenguins: Palmer Archipelago (Antarctica) penguin data. R package version 0.1.0, https://allisonhorst.github.io/palmerpenguins/.
Menze, H B, Kelm, Michael B, Splitthoff, N D, Koethe, Ullrich, Hamprecht, A F (2011). “On oblique random forests.” In Machine Learning and Knowledge Discovery in Databases: European Conference, ECML PKDD 2011, Athens, Greece, September 5-9, 2011, Proceedings, Part II 22, 453-469. Springer. Greenwell, M B, Boehmke, C B, McCarthy, J A (2018). “A simple and effective model-based variable importance measure.” arXiv preprint arXiv:1805.04755.
基于机器学习构建临床预测模型
MachineLearning 2. 因子分析(Factor Analysis)
MachineLearning 3. 聚类分析(Cluster Analysis)
MachineLearning 4. 癌症诊断方法之 K-邻近算法(KNN)
MachineLearning 5. 癌症诊断和分子分型方法之支持向量机(SVM)
MachineLearning 6. 癌症诊断机器学习之分类树(Classification Trees)
MachineLearning 7. 癌症诊断机器学习之回归树(Regression Trees)
MachineLearning 8. 癌症诊断机器学习之随机森林(Random Forest)
MachineLearning 9. 癌症诊断机器学习之梯度提升算法(Gradient Boosting)
MachineLearning 10. 癌症诊断机器学习之神经网络(Neural network)
MachineLearning 11. 机器学习之随机森林生存分析(randomForestSRC)
MachineLearning 12. 机器学习之降维方法t-SNE及可视化(Rtsne)
MachineLearning 13. 机器学习之降维方法UMAP及可视化 (umap)
MachineLearning 14. 机器学习之集成分类器(AdaBoost)
MachineLearning 15. 机器学习之集成分类器(LogitBoost)
MachineLearning 16. 机器学习之梯度提升机(GBM)
MachineLearning 17. 机器学习之围绕中心点划分算法(PAM)
MachineLearning 18. 机器学习之贝叶斯分析类器(Naive Bayes)
MachineLearning 19. 机器学习之神经网络分类器(NNET)
MachineLearning 20. 机器学习之袋装分类回归树(Bagged CART)
MachineLearning 21. 机器学习之临床医学上的生存分析(xgboost)
MachineLearning 22. 机器学习之有监督主成分分析筛选基因(SuperPC)
MachineLearning 23. 机器学习之岭回归预测基因型和表型(Ridge)
MachineLearning 24. 机器学习之似然增强Cox比例风险模型筛选变量及预后估计(CoxBoost)
MachineLearning 25. 机器学习之支持向量机应用于生存分析(survivalsvm)
MachineLearning 26. 机器学习之弹性网络算法应用于生存分析(Enet)
MachineLearning 27. 机器学习之逐步Cox回归筛选变量(StepCox)
MachineLearning 28. 机器学习之偏最小二乘回归应用于生存分析(plsRcox)
MachineLearning 29. 机器学习之嵌套交叉验证(Nested CV)
MachineLearning 30. 机器学习之特征选择森林之神(Boruta)
MachineLearning 31. 机器学习之基于RNA-seq的基因特征筛选 (GeneSelectR)
MachineLearning 32. 机器学习之支持向量机递归特征消除的特征筛选 (mSVM-RFE)
MachineLearning 33. 机器学习之时间-事件预测与神经网络和Cox回归
MachineLearning 34. 机器学习之竞争风险生存分析的深度学习方法(DeepHit)
MachineLearning 35. 机器学习之Lasso+Cox回归筛选变量 (LassoCox)
MachineLearning 36. 机器学习之基于神经网络的Cox比例风险模型 (Deepsurv)
桓峰基因,铸造成功的您!
未来桓峰基因公众号将不间断的推出单细胞系列生信分析教程,
敬请期待!!
桓峰基因官网正式上线,请大家多多关注,还有很多不足之处,大家多多指正!http://www.kyohogene.com/
桓峰基因和投必得合作,文章润色优惠85折,需要文章润色的老师可以直接到网站输入领取桓峰基因专属优惠券码:KYOHOGENE,然后上传,付款时选择桓峰基因优惠券即可享受85折优惠哦!https://www.topeditsci.com/























 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








