安装checkm
刚开始用官网命令 conda install hmmer prodigal pplacer 报错
(vicent) yanziming@server1:~/vicent$ conda install hmmer prodigal pplacer
Collecting package metadata (repodata.json): done
Solving environment: failed with initial frozen solve. Retrying with flexible solve.
PackagesNotFoundError: The following packages are not available from current channels:
- prodigal
- hmmer
- pplacer
Current channels:
- https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/pytorch/linux-64
- https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/pytorch/noarch
- https://repo.anaconda.com/pkgs/main/linux-64
- https://repo.anaconda.com/pkgs/main/noarch
- https://repo.anaconda.com/pkgs/r/linux-64
- https://repo.anaconda.com/pkgs/r/noarch
To search for alternate channels that may provide the conda package you're
looking for, navigate to
https://anaconda.org
and use the search bar at the top of the page.
去官网搜对应的包,选择对应的版本,找到对应的命令,如下图;
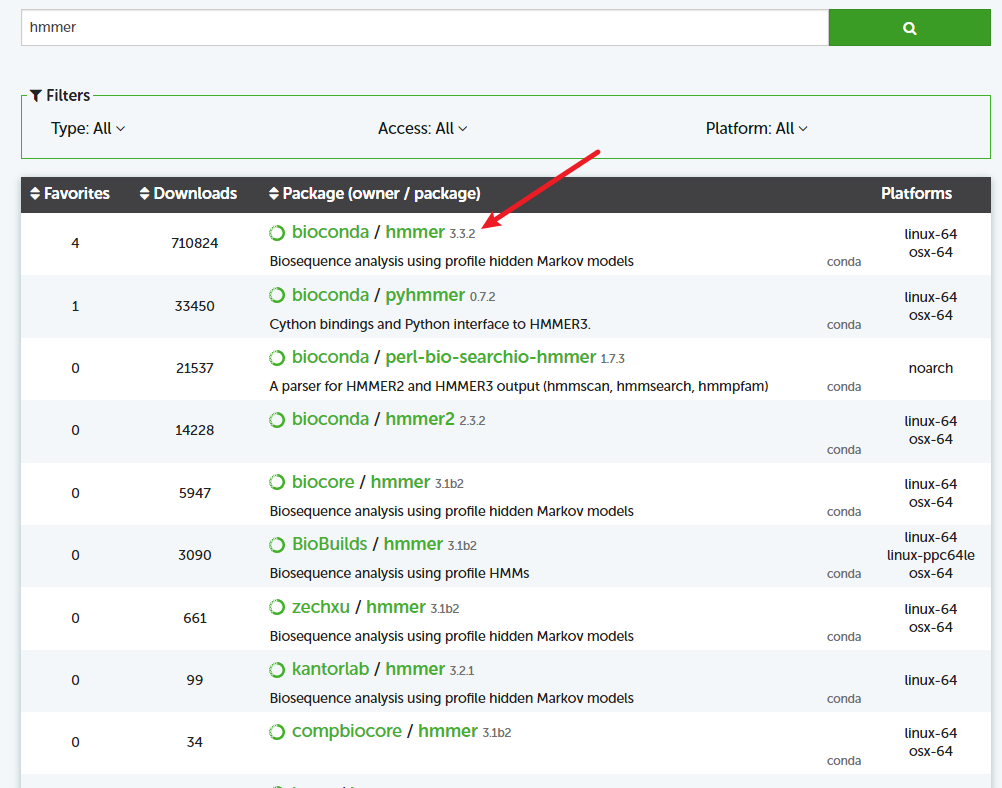
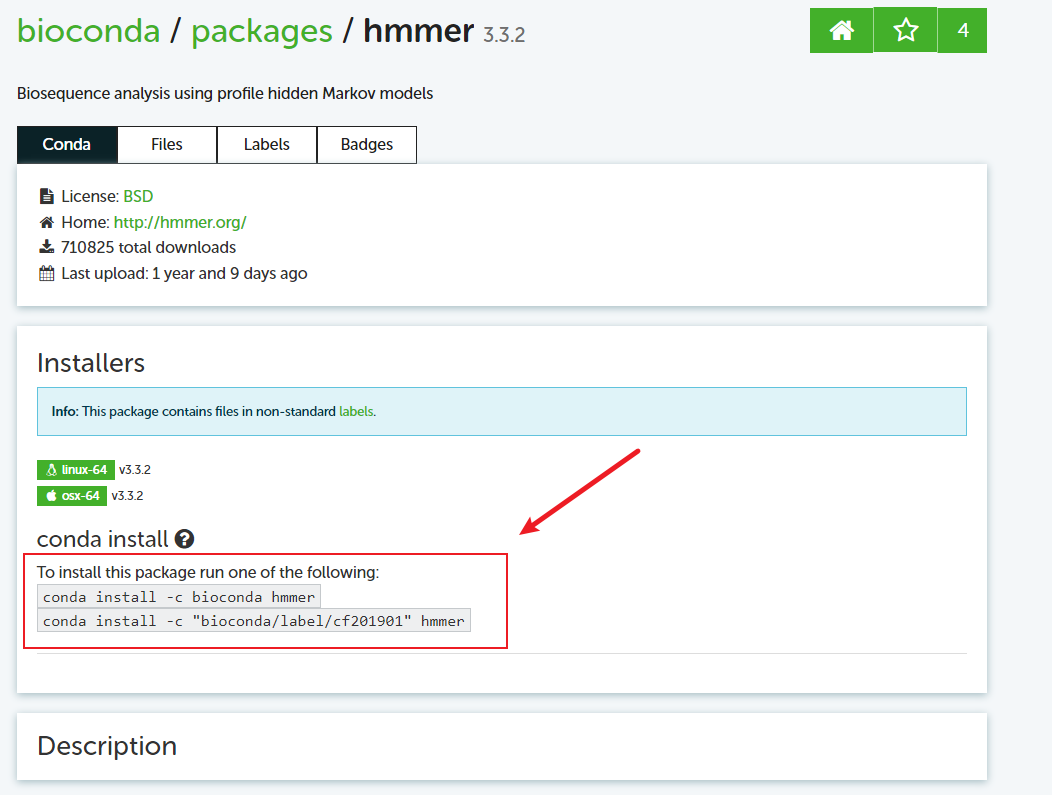
# 安装依赖
conda install -c bioconda prodigal
conda install -c bioconda pplacer
conda install -c bioconda hmmer
# 安装
pip3 install checkm-genome
# 测试
(vicent) yanziming@server1:~/vicent$ checkm
It seems that the CheckM data folder has not been set yet or has been removed. Please run 'checkm data setRoot'.
Path [/home/yanziming/.checkm] does not exist so I will attempt to create it
Path [/home/yanziming/.checkm] has been created and you have permission to write to this folder.
(re) creating manifest file (please be patient).
...::: CheckM v1.2.2 :::...
Lineage-specific marker set:
tree -> Place bins in the reference genome tree
tree_qa -> Assess phylogenetic markers found in each bin
lineage_set -> Infer lineage-specific marker sets for each bin
Taxonomic-specific marker set:
taxon_list -> List available taxonomic-specific marker sets
taxon_set -> Generate taxonomic-specific marker set
Apply marker set to genome bins:
analyze -> Identify marker genes in bins
qa -> Assess bins for contamination and completeness
Common workflows (combines above commands):
lineage_wf -> Runs tree, lineage_set, analyze, qa
taxonomy_wf -> Runs taxon_set, analyze, qa
Reference distribution plots:
gc_plot -> Create GC histogram and delta-GC plot
coding_plot -> Create coding density (CD) histogram and delta-CD plot
tetra_plot -> Create tetranucleotide distance (TD) histogram and delta-TD plot
dist_plot -> Create image with GC, CD, and TD distribution plots together
General plots:
nx_plot -> Create Nx-plots
len_hist -> Sequence length histogram
marker_plot -> Plot position of marker genes on sequences
gc_bias_plot -> Plot bin coverage as a function of GC
Bin exploration and modification:
unique -> Ensure no sequences are assigned to multiple bins
merge -> Identify bins with complementary sets of marker genes
outliers -> [Experimental] Identify outlier in bins relative to reference distributions
modify -> [Experimental] Modify sequences in a bin
Utility functions:
unbinned -> Identify unbinned sequences
coverage -> Calculate coverage of sequences
tetra -> Calculate tetranucleotide signature of sequences
profile -> Calculate percentage of reads mapped to each bin
ssu_finder -> Identify SSU (16S/18S) rRNAs in sequences
Use 'checkm data setRoot <checkm_data_dir>' to specify the location of CheckM database files.
Usage: checkm <command> -h for command specific help
数据下载
数据要不下载之后运行会报文件找不到的错误

# 下载后解压
tar -zxvf checkm_data_2015_01_16.tar.gz
# 运行checkm
checkm lineage_wf -x fasta ./result/ ./result_CHECKM/ -t 30 --tab_table -f test.tab -t 50





















 2728
2728











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








