
当前有两个文件可用:
af-only-gnomad.hg38.vcf.gz (GATK提供)
gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf (gnomAD官网)
将并行处理,最后比较。
# gnomAD官网数据源下载
wget -c https://storage.googleapis.com/gcp-public-data--gnomad/release/2.1.1/liftover_grch38/vcf/exomes/gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf.bgz &看下信息
bcftools view -H af-only-gnomad.hg38.vcf.gz | head -n 3#CHROM POS ID REF ALT QUAL FILTER INFO
chr1 10067 . T TAACCCTAACCCTAACCCTAACCCTAACCCTAACCCTAACCC 30.35 PASS AC=3;AF=7.384e-05
chr1 10108 . CAACCCT C 46514.3 PASS AC=6;AF=0.0001525
chr1 10109 . AACCCT A 89837.3 PASS AC=48;AF=0.001223
# 发现此文件含有多等位基因位点
zcat af-only-gnomad.hg38.vcf.gz | grep -v '##' | grep ',' | head
字段的含义
AC, Alternate allele count for samples
AC0, Allele count is zero after filtering out low-confidence genotypes (GQ < 20; DP < 10; and AB < 0.2 for het calls)
AN, Total number of alleles in samples
AF, Alternate allele frequency in samples
AF_raw, Alternate allele frequency in samples, before removing low-confidence genotypes
AF_eas, Alternate allele frequency in samples of East Asian ancestry
RF, Failed random forest filtering thresholds of 0.055272738028512555, 0.20641025579497013 (probabilities of being a true positive variant) for SNPs, indels
PASS, Passed all variant filters
AN如果为0,说明该位点未测到,AF值也就不可信?
bcftools view -H gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | head -n 1#CHROM POS ID REF ALT QUAL FILTER INFO
chr1 12198 rs62635282 G C 9876.24 AC0 AC=0;AC_afr=0;AC_afr_female=0;AC_afr_male=0;AC_amr=0;AC_amr_female=0;AC_amr_male=0;AC_asj=0;AC_asj_female=0;AC_asj_male=0;AC_eas=0;AC_eas_female=0;AC_eas_jpn=0;AC_eas_kor=0;AC_eas_male=0;AC_eas_oea=0;AC_female=0;AC_fin=0;AC_fin_female=0;AC_fin_male=0;AC_male=0;AC_nfe=0;AC_nfe_bgr=0;AC_nfe_est=0;AC_nfe_female=0;AC_nfe_male=0;AC_nfe_nwe=0;AC_nfe_onf=0;AC_nfe_seu=0;AC_nfe_swe=0;AC_oth=0;AC_oth_female=0;AC_oth_male=0;AC_raw=227;AC_sas=0;AC_sas_female=0;AC_sas_male=0;AF_raw=0.0457108;AN=0;AN_afr=0;AN_afr_female=0;AN_afr_male=0;AN_amr=0;AN_amr_female=0;AN_amr_male=0;AN_asj=0;AN_asj_female=0;AN_asj_male=0;AN_eas=0;AN_eas_female=0;AN_eas_jpn=0;AN_eas_kor=0;AN_eas_male=0;AN_eas_oea=0;AN_female=0;AN_fin=0;AN_fin_female=0;AN_fin_male=0;AN_male=0;AN_nfe=0;AN_nfe_bgr=0;AN_nfe_est=0;AN_nfe_female=0;AN_nfe_male=0;AN_nfe_nwe=0;AN_nfe_onf=0;AN_nfe_seu=0;AN_nfe_swe=0;AN_oth=0;AN_oth_female=0;AN_oth_male=0;AN_raw=4966;AN_sas=0;AN_sas_female=0;AN_sas_male=0;BaseQRankSum=0;ClippingRankSum=0.358;DP=9204;FS=0;InbreedingCoeff=0.0098;MQ=23.04;MQRankSum=0.736;OriginalContig=1;OriginalStart=12198;QD=13.95;ReadPosRankSum=0.736;SOR=0.302;VQSLOD=1.01;VQSR_culprit=MQ;ab_hist_alt_bin_freq=0|0|0|0|1|0|2|0|2|0|10|0|1|28|0|3|0|0|0|0;age_hist_het_bin_freq=0|0|0|0|0|0|0|0|0|0;age_hist_het_n_larger=0;age_hist_het_n_smaller=0;age_hist_hom_bin_freq=0|0|0|0|0|0|0|0|0|0;age_hist_hom_n_larger=0;age_hist_hom_n_smaller=0;allele_type=snv;controls_AC=0;controls_AC_afr=0;controls_AC_afr_female=0;controls_AC_afr_male=0;controls_AC_amr=0;controls_AC_amr_female=0;controls_AC_amr_male=0;controls_AC_asj=0;controls_AC_asj_female=0;controls_AC_asj_male=0;controls_AC_eas=0;controls_AC_eas_female=0;controls_AC_eas_jpn=0;controls_AC_eas_kor=0;controls_AC_eas_male=0;controls_AC_eas_oea=0;controls_AC_female=0;controls_AC_fin=0;controls_AC_fin_female=0;controls_AC_fin_male=0;controls_AC_male=0;controls_AC_nfe=0;controls_AC_nfe_bgr=0;controls_AC_nfe_est=0;controls_AC_nfe_female=0;controls_AC_nfe_male=0;controls_AC_nfe_nwe=0;controls_AC_nfe_onf=0;controls_AC_nfe_seu=0;controls_AC_nfe_swe=0;controls_AC_oth=0;controls_AC_oth_female=0;controls_AC_oth_male=0;controls_AC_raw=109;controls_AC_sas=0;controls_AC_sas_female=0;controls_AC_sas_male=0;controls_AF_raw=0.046661;controls_AN=0;controls_AN_afr=0;controls_AN_afr_female=0;controls_AN_afr_male=0;controls_AN_amr=0;controls_AN_amr_female=0;controls_AN_amr_male=0;controls_AN_asj=0;controls_AN_asj_female=0;controls_AN_asj_male=0;controls_AN_eas=0;controls_AN_eas_female=0;controls_AN_eas_jpn=0;controls_AN_eas_kor=0;controls_AN_eas_male=0;controls_AN_eas_oea=0;controls_AN_female=0;controls_AN_fin=0;controls_AN_fin_female=0;controls_AN_fin_male=0;controls_AN_male=0;controls_AN_nfe=0;controls_AN_nfe_bgr=0;controls_AN_nfe_est=0;controls_AN_nfe_female=0;controls_AN_nfe_male=0;controls_AN_nfe_nwe=0;controls_AN_nfe_onf=0;controls_AN_nfe_seu=0;controls_AN_nfe_swe=0;controls_AN_oth=0;controls_AN_oth_female=0;controls_AN_oth_male=0;controls_AN_raw=2336;controls_AN_sas=0;controls_AN_sas_female=0;controls_AN_sas_male=0;controls_faf95=0;controls_faf95_afr=0;controls_faf95_amr=0;controls_faf95_eas=0;controls_faf95_nfe=0;controls_faf95_sas=0;controls_faf99=0;controls_faf99_afr=0;controls_faf99_amr=0;controls_faf99_eas=0;controls_faf99_nfe=0;controls_faf99_sas=0;controls_nhomalt=0;controls_nhomalt_afr=0;controls_nhomalt_afr_female=0;controls_nhomalt_afr_male=0;controls_nhomalt_amr=0;controls_nhomalt_amr_female=0;controls_nhomalt_amr_male=0;controls_nhomalt_asj=0;controls_nhomalt_asj_female=0;controls_nhomalt_asj_male=0;controls_nhomalt_eas=0;controls_nhomalt_eas_female=0;controls_nhomalt_eas_jpn=0;controls_nhomalt_eas_kor=0;controls_nhomalt_eas_male=0;controls_nhomalt_eas_oea=0;controls_nhomalt_female=0;controls_nhomalt_fin=0;controls_nhomalt_fin_female=0;controls_nhomalt_fin_male=0;controls_nhomalt_male=0;controls_nhomalt_nfe=0;controls_nhomalt_nfe_bgr=0;controls_nhomalt_nfe_est=0;controls_nhomalt_nfe_female=0;controls_nhomalt_nfe_male=0;controls_nhomalt_nfe_nwe=0;controls_nhomalt_nfe_onf=0;controls_nhomalt_nfe_seu=0;controls_nhomalt_nfe_swe=0;controls_nhomalt_oth=0;controls_nhomalt_oth_female=0;controls_nhomalt_oth_male=0;controls_nhomalt_raw=44;controls_nhomalt_sas=0;controls_nhomalt_sas_female=0;controls_nhomalt_sas_male=0;dp_hist_all_bin_freq=125724|24|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0;dp_hist_all_n_larger=0;dp_hist_alt_bin_freq=130|7|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0|0;dp_hist_alt_n_larger=0;faf95=0;faf95_afr=0;faf95_amr=0;faf95_eas=0;faf95_nfe=0;faf95_sas=0;faf99=0;faf99_afr=0;faf99_amr=0;faf99_eas=0;faf99_nfe=0;faf99_sas=0;gq_hist_all_bin_freq=1898|511|26|28|8|4|0|5|2|0|0|0|1|0|0|0|0|0|0|0;gq_hist_alt_bin_freq=14|78|1|25|7|4|0|5|2|0|0|0|1|0|0|0|0|0|0|0;n_alt_alleles=1;nhomalt=0;nhomalt_afr=0;nhomalt_afr_female=0;nhomalt_afr_male=0;nhomalt_amr=0;nhomalt_amr_female=0;nhomalt_amr_male=0;nhomalt_asj=0;nhomalt_asj_female=0;nhomalt_asj_male=0;nhomalt_eas=0;nhomalt_eas_female=0;nhomalt_eas_jpn=0;nhomalt_eas_kor=0;nhomalt_eas_male=0;nhomalt_eas_oea=0;nhomalt_female=0;nhomalt_fin=0;nhomalt_fin_female=0;nhomalt_fin_male=0;nhomalt_male=0;nhomalt_nfe=0;nhomalt_nfe_bgr=0;nhomalt_nfe_est=0;nhomalt_nfe_female=0;nhomalt_nfe_male=0;nhomalt_nfe_nwe=0;nhomalt_nfe_onf=0;nhomalt_nfe_seu=0;nhomalt_nfe_swe=0;nhomalt_oth=0;nhomalt_oth_female=0;nhomalt_oth_male=0;nhomalt_raw=90;nhomalt_sas=0;nhomalt_sas_female=0;nhomalt_sas_male=0;non_cancer_AC=0;non_cancer_AC_afr=0;non_cancer_AC_afr_female=0;non_cancer_AC_afr_male=0;non_cancer_AC_amr=0;non_cancer_AC_amr_female=0;non_cancer_AC_amr_male=0;non_cancer_AC_asj=0;non_cancer_AC_asj_female=0;non_cancer_AC_asj_male=0;non_cancer_AC_eas=0;non_cancer_AC_eas_female=0;non_cancer_AC_eas_jpn=0;non_cancer_AC_eas_kor=0;non_cancer_AC_eas_male=0;non_cancer_AC_eas_oea=0;non_cancer_AC_female=0;non_cancer_AC_fin=0;non_cancer_AC_fin_female=0;non_cancer_AC_fin_male=0;non_cancer_AC_male=0;non_cancer_AC_nfe=0;non_cancer_AC_nfe_bgr=0;non_cancer_AC_nfe_est=0;non_cancer_AC_nfe_female=0;non_cancer_AC_nfe_male=0;non_cancer_AC_nfe_nwe=0;non_cancer_AC_nfe_onf=0;non_cancer_AC_nfe_seu=0;non_cancer_AC_nfe_swe=0;non_cancer_AC_oth=0;non_cancer_AC_oth_female=0;non_cancer_AC_oth_male=0;non_cancer_AC_raw=227;non_cancer_AC_sas=0;non_cancer_AC_sas_female=0;non_cancer_AC_sas_male=0;non_cancer_AF_raw=0.0457293;non_cancer_AN=0;non_cancer_AN_afr=0;non_cancer_AN_afr_female=0;non_cancer_AN_afr_male=0;non_cancer_AN_amr=0;non_cancer_AN_amr_female=0;non_cancer_AN_amr_male=0;non_cancer_AN_asj=0;non_cancer_AN_asj_female=0;non_cancer_AN_asj_male=0;non_cancer_AN_eas=0;non_cancer_AN_eas_female=0;non_cancer_AN_eas_jpn=0;non_cancer_AN_eas_kor=0;non_cancer_AN_eas_male=0;non_cancer_AN_eas_oea=0;non_cancer_AN_female=0;non_cancer_AN_fin=0;non_cancer_AN_fin_female=0;non_cancer_AN_fin_male=0;non_cancer_AN_male=0;non_cancer_AN_nfe=0;non_cancer_AN_nfe_bgr=0;non_cancer_AN_nfe_est=0;non_cancer_AN_nfe_female=0;non_cancer_AN_nfe_male=0;non_cancer_AN_nfe_nwe=0;non_cancer_AN_nfe_onf=0;non_cancer_AN_nfe_seu=0;non_cancer_AN_nfe_swe=0;non_cancer_AN_oth=0;non_cancer_AN_oth_female=0;non_cancer_AN_oth_male=0;non_cancer_AN_raw=4964;non_cancer_AN_sas=0;non_cancer_AN_sas_female=0;non_cancer_AN_sas_male=0;non_cancer_faf95=0;non_cancer_faf95_afr=0;non_cancer_faf95_amr=0;non_cancer_faf95_eas=0;non_cancer_faf95_nfe=0;non_cancer_faf95_sas=0;non_cancer_faf99=0;non_cancer_faf99_afr=0;non_cancer_faf99_amr=0;non_cancer_faf99_eas=0;non_cancer_faf99_nfe=0;non_cancer_faf99_sas=0;non_cancer_nhomalt=0;non_cancer_nhomalt_afr=0;non_cancer_nhomalt_afr_female=0;non_cancer_nhomalt_afr_male=0;non_cancer_nhomalt_amr=0;non_cancer_nhomalt_amr_female=0;non_cancer_nhomalt_amr_male=0;non_cancer_nhomalt_asj=0;non_cancer_nhomalt_asj_female=0;non_cancer_nhomalt_asj_male=0;non_cancer_nhomalt_eas=0;non_cancer_nhomalt_eas_female=0;non_cancer_nhomalt_eas_jpn=0;non_cancer_nhomalt_eas_kor=0;non_cancer_nhomalt_eas_male=0;non_cancer_nhomalt_eas_oea=0;non_cancer_nhomalt_female=0;non_cancer_nhomalt_fin=0;non_cancer_nhomalt_fin_female=0;non_cancer_nhomalt_fin_male=0;non_cancer_nhomalt_male=0;non_cancer_nhomalt_nfe=0;non_cancer_nhomalt_nfe_bgr=0;non_cancer_nhomalt_nfe_est=0;non_cancer_nhomalt_nfe_female=0;non_cancer_nhomalt_nfe_male=0;non_cancer_nhomalt_nfe_nwe=0;non_cancer_nhomalt_nfe_onf=0;non_cancer_nhomalt_nfe_seu=0;non_cancer_nhomalt_nfe_swe=0;non_cancer_nhomalt_oth=0;non_cancer_nhomalt_oth_female=0;non_cancer_nhomalt_oth_male=0;non_cancer_nhomalt_raw=90;non_cancer_nhomalt_sas=0;non_cancer_nhomalt_sas_female=0;non_cancer_nhomalt_sas_male=0;non_neuro_AC=0;non_neuro_AC_afr=0;non_neuro_AC_afr_female=0;non_neuro_AC_afr_male=0;non_neuro_AC_amr=0;non_neuro_AC_amr_female=0;non_neuro_AC_amr_male=0;non_neuro_AC_asj=0;non_neuro_AC_asj_female=0;non_neuro_AC_asj_male=0;non_neuro_AC_eas=0;non_neuro_AC_eas_female=0;non_neuro_AC_eas_jpn=0;non_neuro_AC_eas_kor=0;non_neuro_AC_eas_male=0;non_neuro_AC_eas_oea=0;non_neuro_AC_female=0;non_neuro_AC_fin=0;non_neuro_AC_fin_female=0;non_neuro_AC_fin_male=0;non_neuro_AC_male=0;non_neuro_AC_nfe=0;non_neuro_AC_nfe_bgr=0;non_neuro_AC_nfe_est=0;non_neuro_AC_nfe_female=0;non_neuro_AC_nfe_male=0;non_neuro_AC_nfe_nwe=0;non_neuro_AC_nfe_onf=0;non_neuro_AC_nfe_seu=0;non_neuro_AC_nfe_swe=0;non_neuro_AC_oth=0;non_neuro_AC_oth_female=0;non_neuro_AC_oth_male=0;non_neuro_AC_raw=225;non_neuro_AC_sas=0;non_neuro_AC_sas_female=0;non_neuro_AC_sas_male=0;non_neuro_AF_raw=0.0470908;non_neuro_AN=0;non_neuro_AN_afr=0;non_neuro_AN_afr_female=0;non_neuro_AN_afr_male=0;non_neuro_AN_amr=0;non_neuro_AN_amr_female=0;non_neuro_AN_amr_male=0;non_neuro_AN_asj=0;non_neuro_AN_asj_female=0;non_neuro_AN_asj_male=0;non_neuro_AN_eas=0;non_neuro_AN_eas_female=0;non_neuro_AN_eas_jpn=0;non_neuro_AN_eas_kor=0;non_neuro_AN_eas_male=0;non_neuro_AN_eas_oea=0;non_neuro_AN_female=0;non_neuro_AN_fin=0;non_neuro_AN_fin_female=0;non_neuro_AN_fin_male=0;non_neuro_AN_male=0;non_neuro_AN_nfe=0;non_neuro_AN_nfe_bgr=0;non_neuro_AN_nfe_est=0;non_neuro_AN_nfe_female=0;non_neuro_AN_nfe_male=0;non_neuro_AN_nfe_nwe=0;non_neuro_AN_nfe_onf=0;non_neuro_AN_nfe_seu=0;non_neuro_AN_nfe_swe=0;non_neuro_AN_oth=0;non_neuro_AN_oth_female=0;non_neuro_AN_oth_male=0;non_neuro_AN_raw=4778;non_neuro_AN_sas=0;non_neuro_AN_sas_female=0;non_neuro_AN_sas_male=0;non_neuro_faf95=0;non_neuro_faf95_afr=0;non_neuro_faf95_amr=0;non_neuro_faf95_eas=0;non_neuro_faf95_nfe=0;non_neuro_faf95_sas=0;non_neuro_faf99=0;non_neuro_faf99_afr=0;non_neuro_faf99_amr=0;non_neuro_faf99_eas=0;non_neuro_faf99_nfe=0;non_neuro_faf99_sas=0;non_neuro_nhomalt=0;non_neuro_nhomalt_afr=0;non_neuro_nhomalt_afr_female=0;non_neuro_nhomalt_afr_male=0;non_neuro_nhomalt_amr=0;non_neuro_nhomalt_amr_female=0;non_neuro_nhomalt_amr_male=0;non_neuro_nhomalt_asj=0;non_neuro_nhomalt_asj_female=0;non_neuro_nhomalt_asj_male=0;non_neuro_nhomalt_eas=0;non_neuro_nhomalt_eas_female=0;non_neuro_nhomalt_eas_jpn=0;non_neuro_nhomalt_eas_kor=0;non_neuro_nhomalt_eas_male=0;non_neuro_nhomalt_eas_oea=0;non_neuro_nhomalt_female=0;non_neuro_nhomalt_fin=0;non_neuro_nhomalt_fin_female=0;non_neuro_nhomalt_fin_male=0;non_neuro_nhomalt_male=0;non_neuro_nhomalt_nfe=0;non_neuro_nhomalt_nfe_bgr=0;non_neuro_nhomalt_nfe_est=0;non_neuro_nhomalt_nfe_female=0;non_neuro_nhomalt_nfe_male=0;non_neuro_nhomalt_nfe_nwe=0;non_neuro_nhomalt_nfe_onf=0;non_neuro_nhomalt_nfe_seu=0;non_neuro_nhomalt_nfe_swe=0;non_neuro_nhomalt_oth=0;non_neuro_nhomalt_oth_female=0;non_neuro_nhomalt_oth_male=0;non_neuro_nhomalt_raw=89;non_neuro_nhomalt_sas=0;non_neuro_nhomalt_sas_female=0;non_neuro_nhomalt_sas_male=0;non_topmed_AC=0;non_topmed_AC_afr=0;non_topmed_AC_afr_female=0;non_topmed_AC_afr_male=0;non_topmed_AC_amr=0;non_topmed_AC_amr_female=0;non_topmed_AC_amr_male=0;non_topmed_AC_asj=0;non_topmed_AC_asj_female=0;non_topmed_AC_asj_male=0;non_topmed_AC_eas=0;non_topmed_AC_eas_female=0;non_topmed_AC_eas_jpn=0;non_topmed_AC_eas_kor=0;non_topmed_AC_eas_male=0;non_topmed_AC_eas_oea=0;non_topmed_AC_female=0;non_topmed_AC_fin=0;non_topmed_AC_fin_female=0;non_topmed_AC_fin_male=0;non_topmed_AC_male=0;non_topmed_AC_nfe=0;non_topmed_AC_nfe_bgr=0;non_topmed_AC_nfe_est=0;non_topmed_AC_nfe_female=0;non_topmed_AC_nfe_male=0;non_topmed_AC_nfe_nwe=0;non_topmed_AC_nfe_onf=0;non_topmed_AC_nfe_seu=0;non_topmed_AC_nfe_swe=0;non_topmed_AC_oth=0;non_topmed_AC_oth_female=0;non_topmed_AC_oth_male=0;non_topmed_AC_raw=218;non_topmed_AC_sas=0;non_topmed_AC_sas_female=0;non_topmed_AC_sas_male=0;non_topmed_AF_raw=0.0459334;non_topmed_AN=0;non_topmed_AN_afr=0;non_topmed_AN_afr_female=0;non_topmed_AN_afr_male=0;non_topmed_AN_amr=0;non_topmed_AN_amr_female=0;non_topmed_AN_amr_male=0;non_topmed_AN_asj=0;non_topmed_AN_asj_female=0;non_topmed_AN_asj_male=0;non_topmed_AN_eas=0;non_topmed_AN_eas_female=0;non_topmed_AN_eas_jpn=0;non_topmed_AN_eas_kor=0;non_topmed_AN_eas_male=0;non_topmed_AN_eas_oea=0;non_topmed_AN_female=0;non_topmed_AN_fin=0;non_topmed_AN_fin_female=0;non_topmed_AN_fin_male=0;non_topmed_AN_male=0;non_topmed_AN_nfe=0;non_topmed_AN_nfe_bgr=0;non_topmed_AN_nfe_est=0;non_topmed_AN_nfe_female=0;non_topmed_AN_nfe_male=0;non_topmed_AN_nfe_nwe=0;non_topmed_AN_nfe_onf=0;non_topmed_AN_nfe_seu=0;non_topmed_AN_nfe_swe=0;non_topmed_AN_oth=0;non_topmed_AN_oth_female=0;non_topmed_AN_oth_male=0;non_topmed_AN_raw=4746;non_topmed_AN_sas=0;non_topmed_AN_sas_female=0;non_topmed_AN_sas_male=0;non_topmed_faf95=0;non_topmed_faf95_afr=0;non_topmed_faf95_amr=0;non_topmed_faf95_eas=0;non_topmed_faf95_nfe=0;non_topmed_faf95_sas=0;non_topmed_faf99=0;non_topmed_faf99_afr=0;non_topmed_faf99_amr=0;non_topmed_faf99_eas=0;non_topmed_faf99_nfe=0;non_topmed_faf99_sas=0;non_topmed_nhomalt=0;non_topmed_nhomalt_afr=0;non_topmed_nhomalt_afr_female=0;non_topmed_nhomalt_afr_male=0;non_topmed_nhomalt_amr=0;non_topmed_nhomalt_amr_female=0;non_topmed_nhomalt_amr_male=0;non_topmed_nhomalt_asj=0;non_topmed_nhomalt_asj_female=0;non_topmed_nhomalt_asj_male=0;non_topmed_nhomalt_eas=0;non_topmed_nhomalt_eas_female=0;non_topmed_nhomalt_eas_jpn=0;non_topmed_nhomalt_eas_kor=0;non_topmed_nhomalt_eas_male=0;non_topmed_nhomalt_eas_oea=0;non_topmed_nhomalt_female=0;non_topmed_nhomalt_fin=0;non_topmed_nhomalt_fin_female=0;non_topmed_nhomalt_fin_male=0;non_topmed_nhomalt_male=0;non_topmed_nhomalt_nfe=0;non_topmed_nhomalt_nfe_bgr=0;non_topmed_nhomalt_nfe_est=0;non_topmed_nhomalt_nfe_female=0;non_topmed_nhomalt_nfe_male=0;non_topmed_nhomalt_nfe_nwe=0;non_topmed_nhomalt_nfe_onf=0;non_topmed_nhomalt_nfe_seu=0;non_topmed_nhomalt_nfe_swe=0;non_topmed_nhomalt_oth=0;non_topmed_nhomalt_oth_female=0;non_topmed_nhomalt_oth_male=0;non_topmed_nhomalt_raw=87;non_topmed_nhomalt_sas=0;non_topmed_nhomalt_sas_female=0;non_topmed_nhomalt_sas_male=0;pab_max=1;rf_label=FP;rf_negative_label;rf_tp_probability=0.836542;rf_train;segdup;variant_type=snv;vep=C|downstream_gene_variant|MODIFIER|WASH7P|ENSG00000227232|Transcript|ENST00000423562|unprocessed_pseudogene||||||||||rs62635282|1|2165|-1||SNV|1|HGNC|38034||||||||||||||||||||||||||||||||||||||||||,C|downstream_gene_variant|MODIFIER|WASH7P|ENSG00000227232|Transcript|ENST00000438504|unprocessed_pseudogene||||||||||rs62635282|1|2165|-1||SNV|1|HGNC|38034|YES|||||||||||||||||||||||||||||||||||||||||,C|non_coding_transcript_exon_variant&non_coding_transcript_variant|MODIFIER|DDX11L1|ENSG00000223972|Transcript|ENST00000450305|transcribed_unprocessed_pseudogene|2/6||ENST00000450305.2:n.68G>C||68|||||rs62635282|1||1||SNV|1|HGNC|37102||||||||||||||||||||||||||||||||||||||||||,C|non_coding_transcript_exon_variant&non_coding_transcript_variant|MODIFIER|DDX11L1|ENSG00000223972|Transcript|ENST00000456328|processed_transcript|1/3||ENST00000456328.2:n.330G>C||330|||||rs62635282|1||1||SNV|1|HGNC|37102|YES|||||||||||||||||||||||||||||||||||||||||,C|downstream_gene_variant|MODIFIER|WASH7P|ENSG00000227232|Transcript|ENST00000488147|unprocessed_pseudogene||||||||||rs62635282|1|2206|-1||SNV|1|HGNC|38034||||||||||||||||||||||||||||||||||||||||||,C|non_coding_transcript_exon_variant&non_coding_transcript_variant|MODIFIER|DDX11L1|ENSG00000223972|Transcript|ENST00000515242|transcribed_unprocessed_pseudogene|1/3||ENST00000515242.2:n.327G>C||327|||||rs62635282|1||1||SNV|1|HGNC|37102||||||||||||||||||||||||||||||||||||||||||,C|non_coding_transcript_exon_variant&non_coding_transcript_variant|MODIFIER|DDX11L1|ENSG00000223972|Transcript|ENST00000518655|transcribed_unprocessed_pseudogene|1/4||ENST00000518655.2:n.325G>C||325|||||rs62635282|1||1||SNV|1|HGNC|37102||||||||||||||||||||||||||||||||||||||||||,C|downstream_gene_variant|MODIFIER|WASH7P|ENSG00000227232|Transcript|ENST00000538476|unprocessed_pseudogene||||||||||rs62635282|1|2213|-1||SNV|1|HGNC|38034||||||||||||||||||||||||||||||||||||||||||,C|downstream_gene_variant|MODIFIER|WASH7P|ENSG00000227232|Transcript|ENST00000541675|unprocessed_pseudogene||||||||||rs62635282|1|2165|-1||SNV|1|HGNC|38034||||||||||||||||||||||||||||||||||||||||||,C|regulatory_region_variant|MODIFIER|||RegulatoryFeature|ENSR00001576075|CTCF_binding_site||||||||||rs62635282|1||||SNV|1||||||||||||||||||||||||||||||||||||||||||||
bcftools annotate -x ^INFO/AC,^INFO/AN,^INFO/AF,^INFO/AF_raw,^INFO/AF_eas gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | \
grep -v "^##" | head -n 20#CHROM POS ID REF ALT QUAL FILTER INFO
chr1 12198 rs62635282 G C 9876.24 AC0 AC=0;AF_raw=0.0457108;AN=0
chr1 12237 rs1324090652 G A 81.96 AC0 AC=0;AF_raw=0.000440995;AN=0
chr1 12259 rs1330604035 G C 37.42 AC0 AC=0;AF=0;AF_raw=0.000155788;AN=2
chr1 12266 rs1442951560 G A 2721.48 AC0 AC=0;AF_raw=0.00434708;AN=0
chr1 12272 rs1281272113 G A 2707.42 AC0 AC=0;AF=0;AF_raw=0.00430126;AN=2
chr1 12554 rs1371050997 A G 68.11 AC0;RF AC=0;AF=0;AF_eas=0;AF_raw=0.000294357;AN=3038
chr1 12559 rs1223049744 G A 1666.64 RF AC=14;AF=0.00472654;AF_eas=0.00259067;AF_raw=0.00287838;AN=2962
chr1 12573 rs1273605438 T C 366.59 PASS AC=2;AF=0.000476644;AF_eas=0;AF_raw=0.000172585;AN=4196
chr1 12586 rs1336625132 C T 223.87 PASS AC=2;AF=0.000558971;AF_eas=0.00220264;AF_raw=6.50675e-05;AN=3578
chr1 12596 rs1211439372 C A 44.76 AC0 AC=0;AF=0;AF_eas=0;AF_raw=2.17855e-05;AN=2952
chr1 12597 rs1272077481 T C 569.92 RF AC=8;AF=0.00275103;AF_eas=0;AF_raw=0.0010003;AN=2908
chr1 12599 rs1437963543 CT C 448.69 AC0 AC=0;AF=0;AF_eas=0;AF_raw=2.18036e-05;AN=2830
chr1 12612 rs1205998786 GGT G 41.94 AC0;RF AC=0;AF=0;AF_eas=0;AF_raw=4.02609e-05;AN=5600
chr1 12625 rs1235144565 G A 55.63 PASS AC=1;AF=0.000174825;AF_eas=0;AF_raw=5.98205e-05;AN=5720
chr1 12659 rs1469036210 G C 3242.59 RF AC=7;AF=0.00106093;AF_eas=0.00394737;AF_raw=0.001232;AN=6598
chr1 12670 rs1182032602 G C 2475.63 RF AC=20;AF=0.00291971;AF_eas=0.00791557;AF_raw=0.00295915;AN=6850
chr1 12672 rs1419072050 C T 3690.4 RF AC=13;AF=0.00200803;AF_eas=0.00531915;AF_raw=0.00194156;AN=6474
chr1 12673 rs1476353024 G A 1057.24 RF AC=10;AF=0.0014339;AF_eas=0;AF_raw=0.0019007;AN=6974
chr1 12680 rs1163072234 G A 796.7 PASS AC=6;AF=0.000839396;AF_eas=0.00683371;AF_raw=0.000667608;AN=7148
bcftools annotate -x ^INFO/AC,^INFO/AN,^INFO/AF,^INFO/AF_raw gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | grep -v "^##" | head -n 20#CHROM POS ID REF ALT QUAL FILTER INFO
chr1 12198 rs62635282 G C 9876.24 AC0 AC=0;AF_raw=0.0457108;AN=0
chr1 12237 rs1324090652 G A 81.96 AC0 AC=0;AF_raw=0.000440995;AN=0
chr1 12259 rs1330604035 G C 37.42 AC0 AC=0;AF=0;AF_raw=0.000155788;AN=2
chr1 12266 rs1442951560 G A 2721.48 AC0 AC=0;AF_raw=0.00434708;AN=0
chr1 12272 rs1281272113 G A 2707.42 AC0 AC=0;AF=0;AF_raw=0.00430126;AN=2
chr1 12554 rs1371050997 A G 68.11 AC0;RF AC=0;AF=0;AF_raw=0.000294357;AN=3038
chr1 12559 rs1223049744 G A 1666.64 RF AC=14;AF=0.00472654;AF_raw=0.00287838;AN=2962
chr1 12573 rs1273605438 T C 366.59 PASS AC=2;AF=0.000476644;AF_raw=0.000172585;AN=4196
chr1 12586 rs1336625132 C T 223.87 PASS AC=2;AF=0.000558971;AF_raw=6.50675e-05;AN=3578
chr1 12596 rs1211439372 C A 44.76 AC0 AC=0;AF=0;AF_raw=2.17855e-05;AN=2952
chr1 12597 rs1272077481 T C 569.92 RF AC=8;AF=0.00275103;AF_raw=0.0010003;AN=2908
chr1 12599 rs1437963543 CT C 448.69 AC0 AC=0;AF=0;AF_raw=2.18036e-05;AN=2830
chr1 12612 rs1205998786 GGT G 41.94 AC0;RF AC=0;AF=0;AF_raw=4.02609e-05;AN=5600
chr1 12625 rs1235144565 G A 55.63 PASS AC=1;AF=0.000174825;AF_raw=5.98205e-05;AN=5720
chr1 12659 rs1469036210 G C 3242.59 RF AC=7;AF=0.00106093;AF_raw=0.001232;AN=6598
chr1 12670 rs1182032602 G C 2475.63 RF AC=20;AF=0.00291971;AF_raw=0.00295915;AN=6850
chr1 12672 rs1419072050 C T 3690.4 RF AC=13;AF=0.00200803;AF_raw=0.00194156;AN=6474
chr1 12673 rs1476353024 G A 1057.24 RF AC=10;AF=0.0014339;AF_raw=0.0019007;AN=6974
chr1 12680 rs1163072234 G A 796.7 PASS AC=6;AF=0.000839396;AF_raw=0.000667608;AN=7148
bcftools annotate -x ^INFO/AC,^INFO/AN,^INFO/AF gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | grep -v "^##" | head -n 20 | sed 's/\t/ /g'#CHROM POS ID REF ALT QUAL FILTER INFO
chr1 12198 rs62635282 G C 9876.24 AC0 AC=0;AN=0
chr1 12237 rs1324090652 G A 81.96 AC0 AC=0;AN=0
chr1 12259 rs1330604035 G C 37.42 AC0 AC=0;AF=0;AN=2
chr1 12266 rs1442951560 G A 2721.48 AC0 AC=0;AN=0
chr1 12272 rs1281272113 G A 2707.42 AC0 AC=0;AF=0;AN=2
chr1 12554 rs1371050997 A G 68.11 AC0;RF AC=0;AF=0;AN=3038
chr1 12559 rs1223049744 G A 1666.64 RF AC=14;AF=0.00472654;AN=2962
chr1 12573 rs1273605438 T C 366.59 PASS AC=2;AF=0.000476644;AN=4196
chr1 12586 rs1336625132 C T 223.87 PASS AC=2;AF=0.000558971;AN=3578
chr1 12596 rs1211439372 C A 44.76 AC0 AC=0;AF=0;AN=2952
chr1 12597 rs1272077481 T C 569.92 RF AC=8;AF=0.00275103;AN=2908
chr1 12599 rs1437963543 CT C 448.69 AC0 AC=0;AF=0;AN=2830
chr1 12612 rs1205998786 GGT G 41.94 AC0;RF AC=0;AF=0;AN=5600
chr1 12625 rs1235144565 G A 55.63 PASS AC=1;AF=0.000174825;AN=5720
chr1 12659 rs1469036210 G C 3242.59 RF AC=7;AF=0.00106093;AN=6598
chr1 12670 rs1182032602 G C 2475.63 RF AC=20;AF=0.00291971;AN=6850
chr1 12672 rs1419072050 C T 3690.4 RF AC=13;AF=0.00200803;AN=6474
chr1 12673 rs1476353024 G A 1057.24 RF AC=10;AF=0.0014339;AN=6974
chr1 12680 rs1163072234 G A 796.7 PASS AC=6;AF=0.000839396;AN=7148
bcftools annotate -x ^INFO/AF gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | bcftools norm -f /db/gatk/hg38/Homo_sapiens_assembly38.fasta --multiallelics -both | grep -v "^##" | cut -f 1,2,3,4,5,8 | sed 's/AF=//g' > gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.txt# 查看结果:
grep -w rs777038595 gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.txtchr1 13417 rs777038595 C A 1.49898e-05
chr1 13417 rs777038595 C CGAGA 0.112528
chr1 13417 rs777038595 C CGGGA 0
chr1 13417 rs777038595 C T 1.49898e-05
数据库文件对多等位基因位点似乎已经拆分完毕:
bcftools annotate -x ^INFO/AF gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | head -n 5000 | grep -w rs777038595chr1 13417 rs777038595 C A 2.63878e+07 PASS AF=1.49898e-05
chr1 13417 rs777038595 C CGAGA 2.63878e+07 PASS AF=0.112528
chr1 13417 rs777038595 C CGGGA 2.63878e+07 AC0 AF=0
chr1 13417 rs777038595 C T 2.63878e+07 PASS AF=1.49898e-05
GATK来源的gnomAD的数据的处理
af-only-gnomad.hg38 (含多等位基因位点)
bcftools annotate -x ^INFO/AF af-only-gnomad.hg38.vcf.gz | bcftools norm -f /db/gatk/hg38/Homo_sapiens_assembly38.fasta --multiallelics -both | grep -v "^##" | cut -f 1,2,3,4,5,8 | sed 's/AF=//g' > af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt

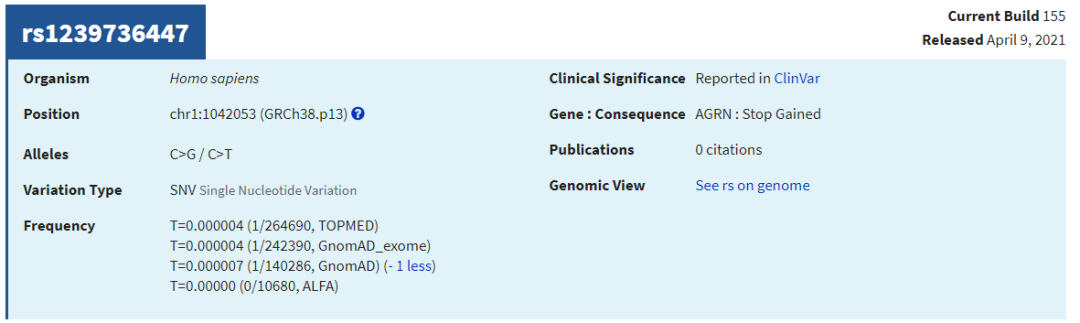
上图dbSNP的"GnomAD_exome"的值来自:
gnomad.exomes.r2.1.1.sites.liftover_grch38

比较:行数
17,201,297
gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.txt
290,331,359
af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt
82,985,813 a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt
比较:标准化历程
Lines total/split/realigned/skipped:17201296/0/6/0
Lines total/split/realigned/skipped:268225276/15895112/9642331/0
比较:内容
af-only-gnomad.hg38,带"chr",含补丁染色体

cat af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | \
grep -P 'chr1\t10140\t'chr1 10140 . ACCCTAAC A 0.0006338
chr1 10140 . A G 0.0001014
多等位基因位点拆分前:
chr1 10140 . ACCCTAAC A,GCCCTAAC 6752.26 PASS AC=25,4;AF=0.0006338,0.0001014
对于gnomad.exomes.r2.1.1
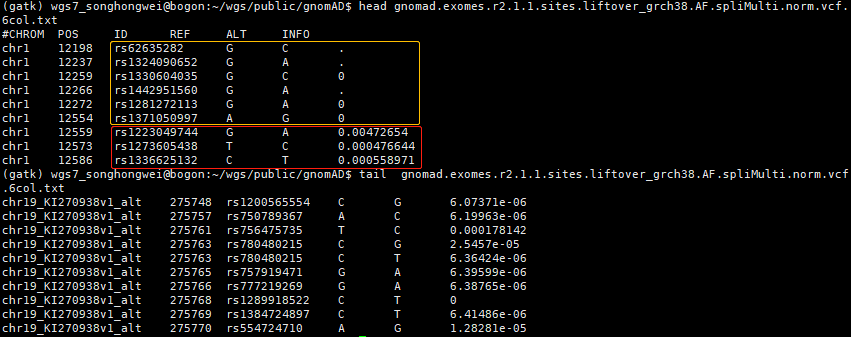
AN, Total number of alleles in samples (AN=0:测序没有测到,或质控后均无基因型)
AC, Alternate allele count for samples
#CHROM POS ID REF ALT QUAL FILTER INFO
chr1 12198 rs62635282 G C 9876.24 AC0 AC=0;AN=0
chr1 12237 rs1324090652 G A 81.96 AC0 AC=0;AN=0
chr1 12259 rs1330604035 G C 37.42 AC0 AC=0;AF=0;AN=2
chr1 12266 rs1442951560 G A 2721.48 AC0 AC=0;AN=0
chr1 12272 rs1281272113 G A 2707.42 AC0 AC=0;AF=0;AN=2
chr1 12554 rs1371050997 A G 68.11 AC0;RF AC=0;AF=0;AN=3038
chr1 12559 rs1223049744 G A 1666.64 RF AC=14;AF=0.00472654;AN=2962
chr1 12573 rs1273605438 T C 366.59 PASS AC=2;AF=0.000476644;AN=4196
chr1 12586 rs1336625132 C T 223.87 PASS AC=2;AF=0.000558971;AN=3578
因此,
gnomad.exomes.r2.1.1文件中,当AF值为 "." 时,测序未测到或样本均无基因型,应该舍弃这些位点;
gnomad.exomes.r2.1.1文件中,值绝对为0的AF,其AC=0,此时的AN值可能很低,也可能很高 (在大样本中确实未发生任何突变),该VCF文件并非"当队列中至少有1个样本变异时,才记录"--这对于AF数据库非常好 (不同于对病人测序样本的处理):有助于增加含有AF值的位点,且这些位点有相当一部分是可信的 (具有较大的AN值)。
af-only-gnomad文件不含 "AC=0"
# af-only-gnomad.hg38 不含 AC=0
bcftools view -H af-only-gnomad.hg38.vcf.gz | \
head -n 20000000 | grep 'AC=0'
# 无返回查看gnomad.exomes.r2.1.1文件中,AC=0时的AN值:
bcftools annotate -x ^INFO/AC,^INFO/AN,^INFO/AF gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | grep -v "^##" | grep 'AC=0' | head -n 200 | cut -f 8 | grep 'AF=0' | grep "AC=\|AF"
虽然AN值很低的情况并不多见,为更严谨一些,最好还是去除这些位点 (例如 AN<10)
nohup bcftools filter -e "AN < 10" -s lowAN gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf | \
bcftools view -H | cut -f 1,2,4,5,7 | \
grep -w lowAN > gnomad.exomes.r2.1.1.sites.liftover_grch38.lowAN.txt &
# 查看
head gnomad.exomes.r2.1.1.sites.liftover_grch38.lowAN.txtchr1 12198 G C lowAN
chr1 12237 G A lowAN
chr1 12259 G C lowAN
chr1 12266 G A lowAN
chr1 12272 G A lowAN
chr1 30524 G A lowAN
chr1 30528 C T lowAN
wc -l gnomad.exomes.r2.1.1.sites.liftover_grch38.lowAN.txt
# 8641
grep -w chr11 gnomad.exomes.r2.1.1.sites.liftover_grch38.lowAN.txt | head
# chr11 400757 G A lowAN
# chr11 627095 C T lowAN
bcftools view -r chr11:400757-400757 gnomad.exomes.r2.1.1.sites.liftover_grch38.vcf.bgz -H
当AF的值不为 "." 且 AN值不低时,输出到文件
nohup awk 'BEGIN{OFS=FS="\t"}ARGIND==1{lowAN["_"$1"_"$2"_"$3"_"$4"_"]=10}ARGIND==2{if($6!="." && lowAN["_"$1"_"$2"_"$4"_"$5"_"]=="") print $0}' \
gnomad.exomes.r2.1.1.sites.liftover_grch38.lowAN.txt \
gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.txt \
> gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt &文件行数:17,192,586
gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt
比较第3个文件源:
gnomad.genomes.r2.1.1.sites.1.liftover_grch38.vcf.bgz (chr1)
来源:

gnomad.genomes的VCF文件由于是全基因组数据,涉及多达1.5万个样本 (无基因型,但INFO列存在大量注释,如不同族群的AF,VEP注释等),导致文件过大,处理起来可能需要好几天。
先只处理chr11,提取其AF,再与现有的文件比较
zcat gnomad.genomes.r2.1.1.sites.1.liftover_grch38.vcf.bgz | bcftools annotate -x ^INFO/AF | grep -v "^##" | cut -f 1,2,3,4,5,8 | sed 's/AF=//g' > gnomad.genomes.r2.1.1.sites.1.liftover_grch38.AF.vcf.6col.txt
zcat af-only-gnomad.hg38.vcf.gz | grep -v "^##" | head -n 4
# zcat gnomad.genomes.r2.1.1.sites.1.liftover_grch38.vcf.bgz | grep -v "##" | head -n 100000 | cut -f 1-5 > test.chr1zcat gnomad.genomes.r2.1.1.sites.1.liftover_grch38.vcf.bgz | grep -v "##" | head -n 100000 | sed 's/AF_raw=/ \t/g' | cut -f 9 | sed 's/;/\t/g' | cut -f 1 > test.af.rawzcat gnomad.genomes.r2.1.1.sites.1.liftover_grch38.vcf.bgz | grep -v "##" | head -n 100000 | sed 's/AF=/ \t/g' | cut -f 9 | sed 's/;/\t/g' | cut -f 1 > test.af
zcat gnomad.genomes.r2.1.1.sites.1.liftover_grch38.vcf.bgz | grep -v "##" | head | grep -w 10109 | grep "AC=\|AF=\|AF_raw=\|AC_raw="
查看几个有rs ID的ClinVar位点,比较AF值
cut -f 10,13 variant_summary_GRCh38.bed.txt | \
nl | grep -v -P '\-1' | grep -w Pathogenic | head


可见,仍有很多致病位点在已发表的大量AF数据库中没有人群频率。因此,筛选致病位点时:1. 可以对AF使用排除法 (排除有AF、且AF值较大的位点);2. 无条件纳入ClinVar/OMIM等已报告的致病位点;3. 当CADD Score极高时也可纳入 (如>50,即使无AF值)。



awk 'BEGIN{OFS=FS="\t"}{if($2<1000000) print $0}' gnomad.genomes.r2.1.1.sites.1.liftover_grch38.AF.vcf.6col.txt > test.gnomad.genomes
head -n 1000000 af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | awk 'BEGIN{OFS=FS="\t"}{if($2<1000000) print $0}' > test.af-only-gnomad
head *test*
tail *test*
比较差异的位点数量
wc -l test.af-only-gnomad test.gnomad.genomes71,842 test.af-only-gnomad
66,454 test.gnomad.genomes
全基因组chr1:1-1,000,000中,含有AF值的总变异数目约7万 (~7%),前者多出5,388。
各自特有的位点:
awk 'BEGIN{OFS=FS="\t"}ARGIND==1{var["_"$1"_"$2"_"$4"_"$5"_"]=1}ARGIND==2{if(var["_"$1"_"$2"_"$4"_"$5"_"]=="") print $0}' \
test.gnomad.genomes test.af-only-gnomad | wc -l5,594
awk 'BEGIN{OFS=FS="\t"}ARGIND==1{var["_"$1"_"$2"_"$4"_"$5"_"]=1}ARGIND==2{if(var["_"$1"_"$2"_"$4"_"$5"_"]=="") print $0}' \
test.af-only-gnomad test.gnomad.genomes | wc -l190
因此gnomad.genomes.r2.1.1来源的位点比af-only-gnomad少约8%,且文件过大。二者都来自gnomAD的全基因组数据。
可使用af-only-gnomad的全基因组AF,使用gnomad.exomes的全外显子组AF。
最终待合并与使用的文件名
af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt
gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt
共3.1亿个短变异的人群频率
往期精品(点击图片直达文字对应教程)
后台回复“生信宝典福利第一波”或点击阅读原文获取教程合集




















































 1007
1007

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








