(2022-09-21)已更正、更新完毕,本篇仅作最后的归档
为什么有关人群频率的推文总是写一篇、再更新一篇?
1. 每个库的文件太大,初次测试好的程序经常要运行数个小时后才能看到结果;2. 如果第2天发现结果文件存在中断或其它报错 (即使问题不是很大),便需要更正、更新文档;3. 人群频率库的"多"和"准"对海量遗传变异的筛选非常重要,需要很小心地求证。
因此每个库的文档都以更新后的为准。按照这些文档,每隔1~2年便更新一次,调用最新的数据库,使分析流程保持在研究的前沿。
gnomAD
ls -sh gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt8.8G (WGS)
gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt
689M (WES)
gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt
1000G
ls -sh a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt3.3G (WGS)
a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt
ALFA
ls -sh AlleleFrequencyAggregator_ALFA/ftp.ensembl/*6col.chr* | cat2.4G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr1.txt
2.6G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr2.txt
2.1G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr3.txt
2.0G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr4.txt
1.9G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr5.txt
1.8G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr6.txt
1.7G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr7.txt
1.6G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr8.txt
1.4G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr9.txt
1.5G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr10.txt
1.5G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr11.txt
1.5G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr12.txt
1.1G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr13.txt
965M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr14.txt
906M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr15.txt
1005M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr16.txt
876M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr17.txt
857M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr18.txt
661M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr19.txt
693M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr20.txt
418M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr21.txt
439M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chr22.txt
1.2G dbSNP154_ALFA_GRCh38....norm.vcf.6col.chrX.txt
41M dbSNP154_ALFA_GRCh38....norm.vcf.6col.chrY.txt
124K dbSNP154_ALFA_GRCh38....norm.vcf.6col.chrM.txt
查看开头、结尾
head -n 3 a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt
head AlleleFrequencyAggregator_ALFA/ftp.ensembl/dbSNP154_ALFA_GRCh38_20210727.Global.spliMulti.norm.vcf.6col.chr1.txt
dbSNP154_ALFA_GRCh38_20210727.Global.spliMulti.norm.vcf.6col.chr1.txt

tail -n 3 a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt
查看列的数目是否一致
nohup cat a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | \
awk 'BEGIN{OFS=FS="\t"}{if(NF!=6) print NR, $0}' \
> wrong.Ncol &合并,且去除重复的短变异
优先次序:gnomAD-exome, ALFA,1000G,gnomAD-genome
# 高频突变 (AF>0.1)
nohup cat gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt \
AlleleFrequencyAggregator_ALFA/ftp.ensembl/*6col.chr* \
a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt \
gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | \
awk 'BEGIN{OFS=FS="\t"}{if(var["_"$1"_"$2"_"$4"_"$5"_"]=="" && $6~/^[0-9.e-]+$/ && $6>0.1) print $1,$2,$3,$4,$5,".",".",$6; var["_"$1"_"$2"_"$4"_"$5"_"]=1}' \
> gnomAD.1000G.AF.moreThan-0.1.txt &
# 文件中断,故拆分成2步,且似乎运行地更快
nohup cat gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt AlleleFrequencyAggregator_ALFA/ftp.ensembl/*6col.chr* a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | awk 'BEGIN{OFS=FS="\t"}{if($6~/^[0-9.e-]+$/ && $6>0.1) print $0}' \
> gnomAD.1000G.AF.moreThan-0.1.redundancy.txt &
awk 'BEGIN{OFS=FS="\t"}{if(var["_"$1"_"$2"_"$4"_"$5"_"]=="") print $1,$2,$3,$4,$5,".",".",$6; var["_"$1"_"$2"_"$4"_"$5"_"]=1}' gnomAD.1000G.AF.moreThan-0.1.redundancy.txt \
> gnomAD.1000G.AF.moreThan-0.1.txt
# 冗余比例约 2.38
wc -l gnomAD.1000G.AF.moreThan-0.1.redundancy.txt gnomAD.1000G.AF.moreThan-0.1.txt
# 21522049 vs. 9053323
# 2.38
# 低频突变 (AF<=0.1)
nohup cat gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt \
AlleleFrequencyAggregator_ALFA/ftp.ensembl/*6col.chr* \
a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt \
gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | \
awk 'BEGIN{OFS=FS="\t"}{if(var["_"$1"_"$2"_"$4"_"$5"_"]=="" && $6~/^[0-9.e-]+$/ && $6<=0.1) print $1,$2,".",$3,$4,$5,".",".",$6; var["_"$1"_"$2"_"$4"_"$5"_"]=1}' \
> gnomAD.1000G.AF.lessThan-0.1.bed &
# 总体冗余比例约 1.32
# 1,420,938,999 vs. 1,078,644,554
# 1.32查看AF值是否都是"数字 (包含如1e-5)"时,注意下面2种用法的区别:

存在重复的变异:
cat a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | awk 'BEGIN{OFS=FS="\t"}{if(var["_"$1"_"$2"_"$4"_"$5"_"]!="") print $0; var["_"$1"_"$2"_"$4"_"$5"_"]=1}' | headchr1 7623266 rs554779128 T TTGAA 0.00405022
chr1 11348703 rs548517482;rs112877363 CTATG C 0.152356
chr1 12924445 rs113511794 C G 0.0896565
chr1 12938980 rs9442277 G A 0.453075
chr1 12941676 rs552423407 C G 0.000399361
chr1 12941687 rs28716506 C T 0.0509185
chr1 12954322 rs7528913 T A 0.973043
chr1 13099405 rs28649915 A T 0.127196
chr1 13099416 rs28565436 A T 0.999601
chr1 13135337 rs1966167 G C 0.465056
cat a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | grep -P 'chr1\t7623266\t'chr1 7623266 rs111828628 T TTGAA 0.985966
chr1 7623266 rs554779128 T TTGAA 0.00405022
chr1 7623266 . T TTGAA 0.001058
chr1 7623266 . TTGAA T 0.008344
chr1 7623266 . TTGAATGAA T 0.0002708
chr1 7623266 . T TTGAATGAA 4.923e-05
cat a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | grep -P 'chr1\t11348703\t'chr1 11348703 rs112877363 CTATG C 0.00119808
chr1 11348703 rs548517482;rs112877363 CTATG C 0.152356
cat a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | grep -P 'chr1\t12924445\t'chr1 12924445 rs113511794 C G 0.838858
chr1 12924445 rs113511794 C G 0.0896565

cat a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt \
gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt \
gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | \
grep -P 'chr1\t12938980\t'chr1 12938980 rs9442277 G A 0.000199681
chr1 12938980 rs9442277 G A 0.453075

cat a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt \
gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt \
gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | \
grep -P 'chr1\t12941687\t'chr1 12941687 rs28716506 C T 0.00419329
chr1 12941687 rs28716506 C T 0.0509185

grep -w rs334 gnomAD.1000G.AF.20220906.txtchr11 5227002 rs334 T A 0.0273562
似乎没有太强的规律。可以调换合并的优先次序,或者取平均值等。或合并、模拟生成bed文件,再用bedtools操作 (排序、索引、去重复、取均值或多数值,等等)。
查看有无遗漏的位点
测试冗余位点中,是否存在既不是高频、也不是低频的位点
# 全部位点
cat gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt \
AlleleFrequencyAggregator_ALFA/ftp.ensembl/*6col.chr* \
a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt \
gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | cut -f 1,2,4,5 > test1
cut -f 1,2,4,5 gnomAD.1000G.AF.moreThan-0.1.txt > test2.more
cut -f 1,2,5,6 gnomAD.1000G.AF.lessThan-0.1.bed > test2.less
cat test2.more test2.less > test2
nohup awk 'BEGIN{OFS=FS="\t"}ARGIND==1{var["_"$1"_"$2"_"$3"_"$4"_"]=1}ARGIND==2{if(var["_"$1"_"$2"_"$3"_"$4"_"]=="") print $0}' \
test1 test2 > loss.site &
# 某个染色体,如:chr2
cat gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt \
AlleleFrequencyAggregator_ALFA/ftp.ensembl/*6col.chr20* \
a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt \
gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | cut -f 1,2,4,5 | \
grep "^chr20" > test1
cut -f 1,2,4,5 gnomAD.1000G.AF.moreThan-0.1.txt | \
grep "^chr20" > test2.more &
cut -f 1,2,5,6 gnomAD.1000G.AF.lessThan-0.1.bed | \
grep "^chr20" > test2.less &
cat test2.more test2.less > test2
nohup awk 'BEGIN{OFS=FS="\t"}ARGIND==1{var["_"$1"_"$2"_"$3"_"$4"_"]=1}ARGIND==2{if(var["_"$1"_"$2"_"$3"_"$4"_"]=="") print $0}' \
test1 test2 > lost.site &无遗漏
再提供一个高、低频AF合并后的文件,这里模拟VCF的前5列,以及AF列,也方便与样本队列的VCF取子集后提交给CADD。
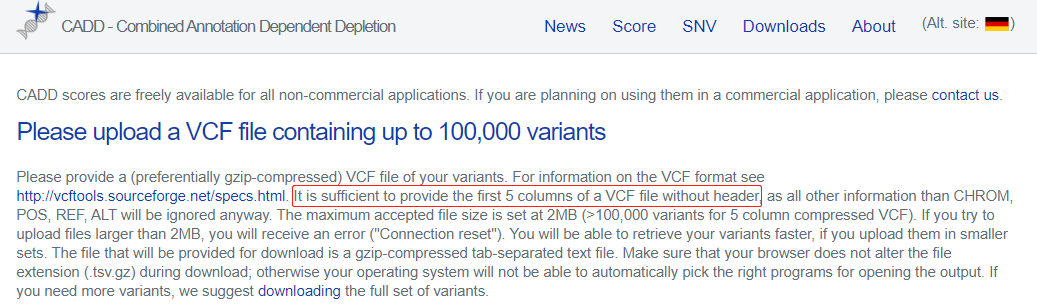
cut -f 1-5,8 gnomAD.1000G.AF.moreThan-0.1.txt > more
cut -f 1,2,4-6,9 gnomAD.1000G.AF.lessThan-0.1.bed > less
cat more less > gnomAD.1000G.ALFA.AF.txt
# 测试某条染色体
cut -f 1,2,4,5 gnomAD.1000G.ALFA.AF.txt | grep "^chrY" > test2
cat gnomAD/gnomad.exomes.r2.1.1.sites.liftover_grch38.AF.spliMulti.norm.vcf.6col.exLowAN.txt AlleleFrequencyAggregator_ALFA/ftp.ensembl/*6col.chrY* \
a1000G/ftp.ensembl/chr/ALL.GRCh38.genotypes.20170504.AF.1samp.spliMulti.norm.vcf.6col.txt gnomAD/af-only-gnomad.hg38.AF.spliMulti.norm.vcf.6col.txt | cut -f 1,2,4,5 | \
grep "^chrY" > test1
# 测试,提交CADD (注意后缀名)
head -n 500 gnomAD.1000G.ALFA.AF.txt > testCADD.vcf

由于WGS的人群频率库很大 (几十亿个位点),须分染色体运行,否则占用内存极大 (>200GB)
# grep -w chr1 gnomAD.1000G.ALFA.AF.txt > gnomAD.1000G.ALFA.AF.chr1.txt &
# grep -w chr11 gnomAD.1000G.ALFA.AF.txt > gnomAD.1000G.ALFA.AF.chr11.txt &
cat Assembly.ChromosomeAccession.Chromosome.25.txt | cut -f 2 | while read chr
do
grep -w ${chr} gnomAD.1000G.ALFA.AF.txt > gnomAD.1000G.ALFA.AF.${chr}.txt &
done
# Check文件大小
ls -shtr gnomAD.1000G.ALFA.AF.chr*.txt
# Check文件行数
wc -l gnomAD.1000G.ALFA.AF.chr*.txt

最终的文件
9,053,323 (高频,约905万)
gnomAD.1000G.AF.moreThan-0.1.txt
1,069,591,231 (低频,约11亿)
gnomAD.1000G.AF.lessThan-0.1.bed
1,078,644,554 (高频 + 低频;含补丁染色体,如chr19_KI270938v1_alt)
gnomAD.1000G.ALFA.AF.txt
gnomAD.1000G.ALFA.AF.chr*.txt (chr1...22XYM)
旧服务器 (1-6期学员) ,已更新至
/home/public/db/
新服务器,已更新至
/disk2/home/shw/db/ (软链接)
/disk4/home/shw/db/
dbSNP库的155版的AF库更大,待后续合并
对每个AF单独处理、再合并的好处是:
可随时更新,以及控制中间过程 (比如选择东亚人种的AF)

往期精品(点击图片直达文字对应教程)
后台回复“生信宝典福利第一波”或点击阅读原文获取教程合集









 文章描述了对多个遗传变异数据库(gnomAD,1000G,ALFA,dbSNP)进行整合和分析的过程,特别是针对高频和低频突变的筛选,以及如何处理文件的冗余和确保数据准确性。作者强调了频繁更新文档的重要性,以反映最新的研究进展,并提供了用于筛选和合并数据的脚本示例。
文章描述了对多个遗传变异数据库(gnomAD,1000G,ALFA,dbSNP)进行整合和分析的过程,特别是针对高频和低频突变的筛选,以及如何处理文件的冗余和确保数据准确性。作者强调了频繁更新文档的重要性,以反映最新的研究进展,并提供了用于筛选和合并数据的脚本示例。











































 914
914

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








