VolcanoSV
VolcanoSV能够从单分子长reads中准确和稳定的识别二倍体基因组中的结构变异(SV),拥有单染色体模式和WGS模式两种运行模式。
github: https://www.github-zh.com/projects/623191841-volcanosv#single-chromosome-mode
详细用法参考: https://www.github-zh.com/projects/623191841-volcanosv#single-chromosome-mode
1. 下载安装
# 克隆
git clone --recurse-submodules https://github.com/maiziezhoulab/VolcanoSV.git
cd VolcanoSV
sh Install.sh
# 创建conda环境, python版本3.8.3
conda env create -f VolcanoSV/requirement.yaml
conda activate volcanosv
# 设置VolcanoSV所在目录,方便后续脚本调用
path_to_volcanosv=/path/to/VolcanoSV
2. 数据下载
对于单染色体模式,提供了用于Hifi、BamHI和ONT数据的chr10 BAM、contigs和VCF测试文件。可以从Zenodo网站下载: https://zenodo.org/records/10520476
针对输入的BAM数据或ONT数据的结果,则只需将输入BAM文件和参数“dtype”更改为相应的(BAM/ONT)数据类型即可。
# lunix命令行下载, 示例数据使用的hg19参考基因组
wget https://cf.10xgenomics.com/supp/genome/refdata-hg19-2.1.0.tar.gz
tar -xzvf refdata-hg19-2.1.0.tar.gz
3. 单染色体模式
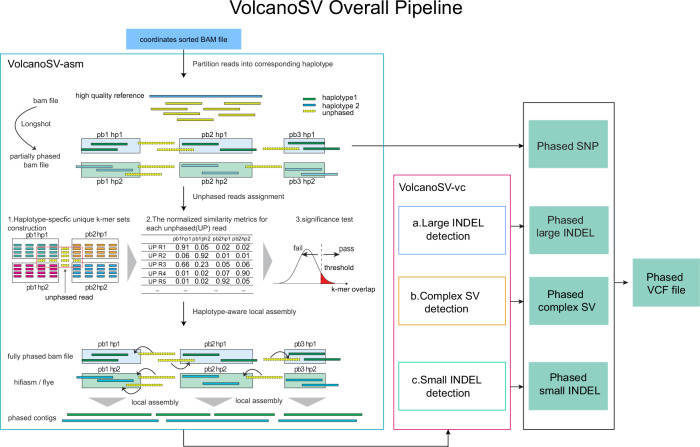
3.1 单染色体模式VolcanoSV组装(VolcanoSV-asm)
VolcanoSV组装流水线被设计为通过染色体运行。我们将多个最先进的汇编器集成到管道中,包括hifiasm,Flye,wtdbg 2,miniasm,Shasta,NextDenovo和Hicanu/Canu。用户可以根据自己的需要选择合适的汇编器。主脚本是${path_to_volcanosv}/bin/VolcanoSV-asm/volcanosv-asm.py。
# 对高保真数据使用hifiasm
# 默认情况下,VolcanoSV对Hifi数据使用hifiasm,对ONT和ONT数据使用Flye
python ${path_to_volcanosv}/bin/VolcanoSV-asm/volcanosv-asm.py \
-bam Hifi_L2_hg19_minimap2_chr10.bam \
-o volcanosv_asm_output \
-ref refdata-hg19-2.1.0/fasta/genome.fa \
-t 10 \
-chr 10 \
-dtype Hifi \
-px Hifi_L2 \
-asm hifiasm
参数说明:
–bam_file INBAM, -bam INBAM, could be either wgs bam or single-chromosome bam file
–output_dir output_dir, -o output_dir
–reference REFERENCE, -ref REFERENCE
–n_thread N_THREAD, -t N_THREAD
–chrnum {1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22}, -chr {1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22}
–assembler {wtdbg2,canu,miniasm,shasta,nextdenovo,hifiasm,hicanu,flye}, -asm {wtdbg2,canu,miniasm,shasta,nextdenovo,hifiasm,hicanu,flye}
optional; if not set, VolcanoSV use hifiasm for Hifi data and flye for CLR and ONT data by default.
–data_type {CLR,ONT,Hifi}, -dtype {CLR,ONT,Hifi}
–pacbio_subtype {rs,sq}, -pb {rs,sq}
must provide when using wtdbg2 on CLR data (default: None)
–shasta_ont_config {Nanopore-OldGuppy-Sep2020}, -shacon {Nanopore-OldGuppy-Sep2020}
–prefix PREFIX, -px PREFIX
3.2 单染色体模式大插入缺失检测(VolcanoSV-vc)
主脚本${path_to_volcanosv}/bin/VolcanoSV-vc/Large_INDEL/volcanosv-vc-large-indel.py
输入目录应该是volcanoSV-asm的输出目录。脚本兼容单染色体模式或wgs模式:当提供参数“chrnum”时,它将在单染色体模式下执行,否则,它将假设input_dir包含chr1-chr 22并在wgs模式下执行。请注意前缀prefix应该与volcanosv-asm中的设置保持一致。运行上述代码后,您将在<ouput_folder>/volcanosv_large_indel.vcf中输出VCF。
python ${path_to_volcanosv}/bin/VolcanoSV-vc/Large_INDEL/volcanosv-vc-large-indel.py \
-i volcanosv_asm_output/ \
-o volcanosv_large_indel_output/ \
-dtype Hifi \
-bam Hifi_L2_hg19_minimap2_chr10.bam \
-ref refdata-hg19-2.1.0/fasta/genome.fa \
-chr 10 -t 10 \
-px Hifi_L2
参数说明:
–input_dir INPUT_DIR, -i INPUT_DIR
–output_dir OUTPUT_DIR, -o OUTPUT_DIR
–data_type DATA_TYPE, -dtype DATA_TYPE
Hifi;CLR;ONT
–bam_file RBAM_FILE, -bam RBAM_FILE
reads bam file for reads signature extraction
–reference REFERENCE, -ref REFERENCE
wgs reference file
–chrnum {1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22}, -chr {1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22}
–n_thread N_THREAD, -t N_THREAD
–n_thread_align N_THREAD_ALIGN, -ta N_THREAD_ALIGN
–mem_per_thread MEM_PER_THREAD, -mempt MEM_PER_THREAD
Set maximum memory per thread for alignment; suffix K/M/G recognized; default = 768M
–prefix PREFIX, -px PREFIX
3.3 单染色体模式小插入缺失检测(VolcanoSV-vc)
主脚本是${path_to_volcanosv}/bin/VolcanoSV-vc/Small_INDEL/volcanosv-vc-small-indel.py。将在volcanosv_small_indel/Hifi_L2_volcanosv_small_indel.vcf中输出VCV。
python ${path_to_volcanosv}/bin/VolcanoSV-vc/Small_INDEL/volcanosv-vc-small-indel.py \
-i volcanosv_asm_output/ \
-o volcanosv_small_indel \
-bam Hifi_L2_hg19_minimap2_chr10.bam \
-ref refdata-hg19-2.1.0/fasta/genome.fa \
-r chr10 \
-t 30 \
-px Hifi_L2
参数说明:
–input_dir INPUT_DIR, -i INPUT_DIR
–bam_file READ_BAM, -bam READ_BAM
–output_dir OUTPUT_DIR, -o OUTPUT_DIR
–reference REFERENCE, -ref REFERENCE
–bedfile BEDFILE, -bed BEDFILE
optional; a high confidence bed file (default: None)
–region REGION, -r REGION
optional; exmaple: chr21:2000000-2100000 (default: None)
–n_thread N_THREAD, -t N_THREAD
–kmer_size KMER_SIZE, -k KMER_SIZE
–ratio RATIO, -rt RATIO
maximum bad kmer ratio (default: 0.3)
–min_support MIN_SUPPORT, -ms MIN_SUPPORT
maximum support for bad kmer (default: 5)
–prefix PREFIX, -px PREFIX
3.4 单染色体模式SNP检测(VolcanoSV-vc)
采用Longshot的SNP calling结果作为最终结果。成功运行组装管道后,phased的SNP VCF文件:volcanosv_asm_output/<chromosome_name>/phasing_result/_phased.vcf。























 688
688

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










