
简 介
Pfam数据库是一个蛋白质家族的大集合,每个家族都由多个序列比对和隐马尔可夫模型(hmm)表示。蛋白质通常由一个或多个功能区组成,通常称为结构域。不同的结构域组合产生了自然界中发现的各种各样的蛋白质。因此,鉴定发生在蛋白质内部的结构域可以深入了解其的功能。Pfam还生成相关条目的高级分组,称为宗族。宗族是由序列、结构或剖面的相似性联系在一起的Pfam条目的集合。每个条目的数据都是基于UniProt参考蛋白质组,但单个UniProtKB序列的信息仍然可以通过输入蛋白质加入来找到。Pfam全比对可以通过搜索各种数据库获得,要么提供不同的接入(例如所有UniProt和NCBI GI),要么提供不同级别的冗余。
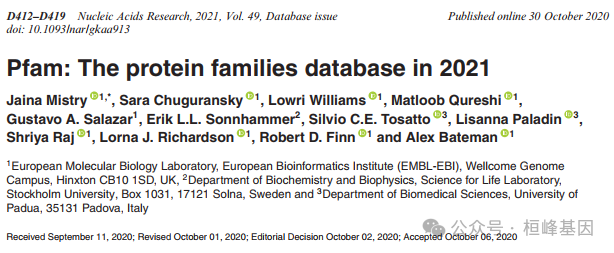
UniProtKB 的增长,以及 Pfam 在过去五个 Pfam 版本中的覆盖范围。随着UniProtKB 大小的增长,Pfam 序列和残基的覆盖率分别保持在~ 77%和~ 53%。图中的 UniProtKB 大小对应于在每个 Pfam 版本中使用的 UniProtKB 版本。
在线分析
最新版本 Pfam 33.1 中增加了350多个新家族,并对现有条目进行了许多改进。为了便于对COVID-19的研究,修订了涵盖SARS-CoV-2蛋白质组的Pfam条目,并为Pfam未涵盖的区域建立了新的条目。重新引入了Pfam-B,提供了Pfam的自动生成补充,包含136730个尚未与Pfam家族匹配的新序列簇。新的Pfam-B是基于MMseqs2软件的聚类。已经将RepeatsDB中的所有区域与Pfam中的区域进行了比较,并开始使用这些结果来构建和完善Pfam重复序列家族(pfam)。

本地分析
数据下载
因为是在服务器上直接操作下载,如果自己的电脑可以自行配置系统,然后进行数据库下载。
Pfam-A为高质量,手工确定的蛋白结构域数据,Pfam-B为基于Pfam-A数据库自动注释得到的蛋白结构域数据库,我们这里只下载了Pfam-A即可。
wget ftp://ftp.ebi.ac.uk:21/pub/databases/Pfam/current_release/Pfam-A.hmm.gz
wget ftp://ftp.ebi.ac.uk:21/pub/databases/Pfam/current_release/Pfam-A.hmm.dat.gz
wget ftp://ftp.ebi.ac.uk:21/pub/databases/Pfam/current_release/active_site.dat.gz
gunzip *.gzhmmer-3.2下载安装
wget http://eddylab.org/software/hmmer/hmmer-3.2.tar.gz
tar -xzvf hmmer-3.2.1.tar.gz
cd hmmer-3.2
./configure
make
make check
make install
# 添加至环境变量
vim ~/.bashrc
export PATH=/usr/local/bin:$PATH
# 环境变量立即生效
source ~/.bashrc新建一个环境安装pfam-scan
默认是已经安装了Anconda/miniconda3,这个需要提取配置好的哦。然后 conda 创建一个新环境,安装pfam-scan,并激活。
conda create -n pfam_scan
source activate pfam_scan
conda install pfam_scan然后利用hmmpress数据库建索引就可以使用了。
hmmpress Pfam-A.hmm测试一下是否安装成功,如下:
pfam_scan.pl -h
pfam_scan.pl: search a FASTA file against a library of Pfam HMMs
Usage: pfam_scan.pl -fasta <fasta_file> -dir <directory location of Pfam files>
Additonal options:
-h : show this help
-outfile <file> : output file, otherwise send to STDOUT
-clan_overlap : show overlapping hits within clan member families (applies to Pfam-A families only)
-align : show the HMM-sequence alignment for each match
-e_seq <n> : specify hmmscan evalue sequence cutoff for Pfam-A searches (default Pfam defined)
-e_dom <n> : specify hmmscan evalue domain cutoff for Pfam-A searches (default Pfam defined)
-b_seq <n> : specify hmmscan bit score sequence cutoff for Pfam-A searches (default Pfam defined)
-b_dom <n> : specify hmmscan bit score domain cutoff for Pfam-A searches (default Pfam defined)
-as : predict active site residues for Pfam-A matches
-json [pretty] : write results in JSON format. If the optional value "pretty" is given,
the JSON output will be formatted using the "pretty" option in the JSON
module
-cpu <n> : number of parallel CPU workers to use for multithreads (default all)
-translate [mode] : treat sequence as DNA and perform six-frame translation before searching. If the
optional value "mode" is given it must be either "all", to translate everything
and produce no individual ORFs, or "orf", to report only ORFs with length greater
than 20. If "-translate" is used without a "mode" value, the default is to
report ORFs (default no translation)
For more help, check the perldoc:
shell% perldoc pfam_scan.pl文件准备
这个输入文件只有一个文件就是蛋白序列文件,例如:
>sp|O95905|ECD_HUMAN Protein ecdysoneless homolog OS=Homo sapiens OX=9606 GN=ECD PE=1 SV=1
MEETMKLATMEDTVEYCLFLIPDESRDSDKHKEILQKYIERIITRFAPMLVPYIWQNQPF
NLKYKPGKGGVPAHMFGVTKFGDNIEDEWFIVYVIKQITKEFPELVARIEDNDGEFLLIE
AADFLPKWLDPENSTNRVFFCHGELCIIPAPRKSGAESWLPTTPPTIPQALNIITAHSEK
ILASESIRAAVNRRIRGYPEKIQASLHRAHCFLPAGIVAVLKQRPRLVAAAVQAFYLRDP
IDLRACRVFKTFLPETRIMTSVTFTKCLYAQLVQQRFVPDRRSGYRLPPPSDPQYRAHEL
GMKLAHGFEILCSKCSPHFSDCKKSLVTASPLWASFLESLKKNDYFKGLIEGSAQYRERL
EMAENYFQLSVDWPESSLAMSPGEEILTLLQTIPFDIEDLKKEAANLPPEDDDQWLDLSP
DQLDQLLQEAVGKKESESVSKEEKEQNYDLTEVSESMKAFISKVSTHKGAELPREPSEAP
ITFDADSFLNYFDKILGPRPNESDSDDLDDEDFECLDSDDDLDFETHEPGEEASLKGTLD
NLKSYMAQMDQELAHTCISKSFTTRNQVEPVSQTTDNNSDEEDSGTGESVMAPVDVDLNL
VSNILESYSSQAGLAGPASNLLQSMGVQLPDNTDHRPTSKPTKN实际操作
pfam_scan.pl -fasta ./test.fa -dir hmmer-3.2/ -outfile test_result.xls -as
#[42] [94] [42] [110] [66] [398] [363] [272] [294] [392] [294] [134] [291] [212] [64] [549][567] [562][580] [515][533] [539][557] [513][531] [513][531] [546][564] [537][555] [550][568] [184] [184] [329] [152] [376][394] [193][211] [293] [273] [614,680] [90] [76] [90] [76] [76] [90] [73] [123] [25] [39] [213] [30] [204] [51] [225] [81] [255] [39] [213] [39] [213] [71] [245] [71] [245] [18,106,126] [18,106,126] [18,106,126] [86] [18,106,126] [67] [210,343] [210,343] [210,343] [85] [184,191] [483,532,633] [173]结果解读
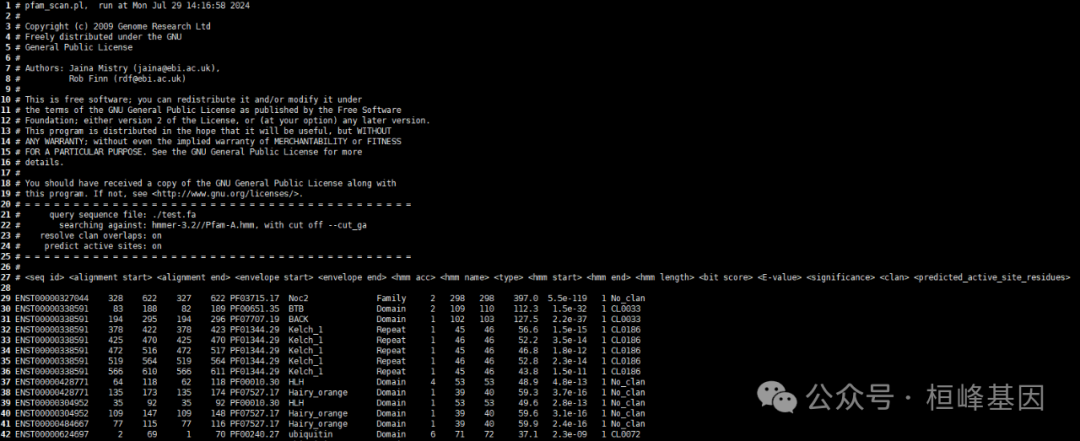
输出结果说明:
(1) seq_id:蛋白序列编号
(2) alignment start:蛋白序列比对的起始位置
(3) alignment end:蛋白序列比对的终止位置
(4) envelope start:蛋白序列结构域的起始位置
(5) envelope end:蛋白序列结构域的终止位置
(6) hmm acc:比对到pfam结构域的ID
(7) hmm name:pfam结构域名称
(8) type:pfam结构域类型
(9) hmm start:比对到结构域的起始位置
(10) hmm end:比对到结构域的终止位置
(11) hmm length:pfam结构域的长度
(12) bit score:比对打分分值
(13) E-value:比对的E值
(14) Significance:比对序列的显著性
(15) Clan:蛋白结构域超级家族名称
(16) predicted_active_site_residues:比对的序列是否位于酶的活性部位
Reference
Pfam: The protein families database in 2021: J. Mistry, et al. Nucleic Acids Research (2021)
桓峰基因,铸造成功的您!
未来桓峰基因公众号将不间断的推出单细胞系列生信分析教程,
敬请期待!!
桓峰基因官网正式上线,请大家多多关注,还有很多不足之处,大家多多指正!http://www.kyohogene.com/
桓峰基因和投必得合作,文章润色优惠85折,需要文章润色的老师可以直接到网站输入领取桓峰基因专属优惠券码:KYOHOGENE,然后上传,付款时选择桓峰基因优惠券即可享受85折优惠哦!https://www.topeditsci.com/






















 4055
4055

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








