一、基本上来看大概由5个评价参数:
- Validity(有效性)
- Uniqueness(独一无二性)
- Novelty(新颖性)
- KL divergence(KL散度)
- FCD(Fréchet ChemNet 距离)
KL-divergence 和 FCD是为了测量生成的分子和训练集、测试集的分子之间的相似性;
validity、FCD、KL-散度 三者的得分高度一致,它们三者的得分与“ novelty ”呈负相关;
二、使用的基准模型为:
- Random sampler: baseline model;
- SMILES LSTM: Long-Short-Term Memory DNN for SMILES strings;
- Graph MCTS: Graph-based Monte Carlo Tree Search;
- AAE: Adversarial AutoEncoder;
- ORGAN: Objective-Reinforced Generative Adversarial Network;
- VAE: Variational AutoEncoder;
- FASMIFRA: Fast Assembly of SMILES Fragments (proposed method);
- Negative control: FASMIFRA without extended bond typing (any fragment can be connected to any other fragment)
1、Molecular De Novo Design through Deep Reinforcement Learning
code:https://github.com/MarcusOlivecrona/REINVENT
paper:https://arxiv.org/abs/1704.07555

Again the consequence of removing the actives from the Prior was a threefold reduction in the probability of generating a test set active: the difference between the two Priors is directly mirrored by their corresponding Agents. Apart from generating a higher fraction of structures that are predicted to be active, both Agents also generate a significantly higher fraction of valid SMILES (Table 4). Sequences that are not valid SMILES receive a score of -1, which means that the scoring function naturally encourages valid SMILES.
2、Masked graph modeling for molecule generation(2021)【nature communications】
paper:Masked graph modeling for molecule generation | Nature Communications
code:GitHub - nyu-dl/dl4chem-mgm
In this work, we evaluate our approach on two popular molecular graph datasets, QM9 、 ChEMBL, using a set of five distribution-learning metrics introduced in the GuacaMol benchmark: the validity, uniqueness, novelty, KL-divergence (KLD) and Fréchet ChemNet Distance (FCD) scores.
(1)、在QM9数据集上使用“ masked graph model ”得出的结果的基准指标之间的Spearman相关系数。
Table 1. Spearman’s correlation coefficient(spearman相关系数) between benchmark metrics for results using the masked graph model on the QM9 dataset.
(2)、在QM9数据集上使用LSTM、Transformer Small和Transformer Regular的结果的基准指标之间的Spearman相关系数。
Table 2. Spearman’s correlation coefficient between benchmark metrics for results using LSTM, Transformer Small and Transformer Regular on the QM9 dataset.
(3)、QM9上的分布结果。CharacterVAE、GrammarVAE、GraphVAE和MolGAN结果取自Cao和Kipf。
Table 3. Distributional results on QM9. CharacterVAE, GrammarVAE, GraphVAE and MolGAN results are taken from Cao and Kipf.

(4)、ChEMBL上的分布结果。LSTM, Graph MCTS, AAE, ORGAN and VAE结果取自Brown等人
Table 4. Distributional results on ChEMBL. LSTM, Graph MCTS, AAE, ORGAN and VAE (with a bidirectional GRU as encoder and autoregressive GRU as decoder) results are taken from Brown et al..

支撑材料:

Supplementary Table 1: Effect of varying masking rate and graph initialization on the benchmark results for our masked graph model on QM9 and ChEMBL.
3、Generative Adversarial Networks for De Novo Molecular Design
paper:https://onlinelibrary.wiley.com/doi/abs/10.1002/minf.202100045

Table 1 summarizes the results of the baseline models and SMILES-MaskGAN for the distribution-learning benchmarks. The baseline models listed in the Table 1 are available at www.github.com/BenevolentAI/guacamol_baselines . The random sampler samples 10,000 molecules from the ChEMBL training dataset with replacement and computes the scores.
The novelty score is zero because it only samples the molecules present in the training data.
Based on the KL divergence score and FCD, the results demonstrate that the SMILES LSTM model reproduces a distribution similar to that of the ChEMBL training dataset. However,
in terms of validity, uniqueness, and novelty, it scored lower than the Graph Monte Carlo Tree Search (Graph MCTS) and the proposed SMILES-MaskGAN.
Although the Graph MCTS model[49] exhibits high scores for validity, uniqueness, and novelty, it fails to reproduce the molecular distribution of the training data. The adversarial autoencoder (AAE) model[50] exhibited good scores for validity, uniqueness, and novelty. However, the distribution of the generated molecules was not close to that of the training data. The variational autoencoder (VAE) model[16] does not have the best score for all benchmarks; however, it yields relatively acceptable scores compared with the other models. In addition, ORGAN,[24] which combines the reinforcement learning method with the GAN architecture, obtains the lowest scores on all benchmarks. Although it has relatively acceptable scores for uniqueness and novelty, it only has a slight similarity with the distribution of training data and does not produce more than half of the valid molecules.
4、Molecular generation by Fast Assembly of (Deep)SMILES fragments【2021】
code:GitHub - UnixJunkie/FASMIFRA: Molecular Generation by Fast Assembly of SMILES Fragments
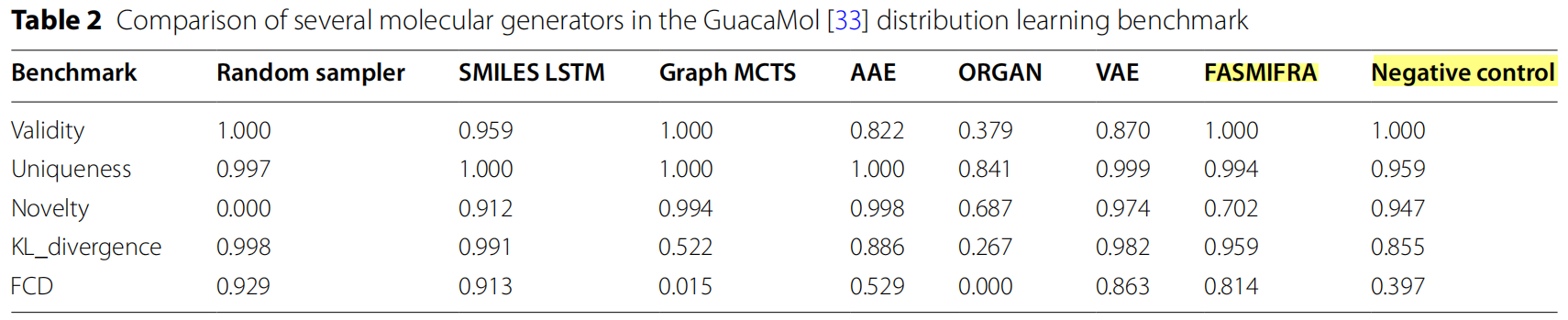
Random sampler: baseline model;
SMILES LSTM: Long-Short-Term Memory DNN for SMILES strings;
Graph MCTS: Graph-based Monte Carlo Tree Search;
AAE: Adversarial AutoEncoder;
ORGAN: Objective-Reinforced Generative Adversarial Network;
VAE: Variational AutoEncoder;
FASMIFRA: Fast Assembly of SMILES Fragments (proposed method);
Negative control: FASMIFRA without extended bond typing (any fragment can be connected to any other fragment)








 本文综述了分子生成领域的评估标准及多种基准模型的表现。通过五个关键指标:有效性、独一无二性、新颖性、KL散度和Fréchet ChemNet距离,评估不同模型在分子生成任务上的性能。文中还对比了多种模型,如SMILES-LSTM、GraphMCTS等,在QM9和ChEMBL数据集上的表现。
本文综述了分子生成领域的评估标准及多种基准模型的表现。通过五个关键指标:有效性、独一无二性、新颖性、KL散度和Fréchet ChemNet距离,评估不同模型在分子生成任务上的性能。文中还对比了多种模型,如SMILES-LSTM、GraphMCTS等,在QM9和ChEMBL数据集上的表现。



















 2320
2320

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










