Bohnsack, Katrin Sophie, et al. "Alignment-free sequence comparison: A systematic survey from a machine learning perspective." IEEE/ACM Transactions on Computational Biology and Bioinformatics 20.1 (2022): 119-135. https://ieeexplore.ieee.org/abstract/document/9672674
- 被引次数:10;
- 期刊:影响因子 3.702;JCR Q1;中科院大类(工程技术)3区,小类(数学跨学科应用、计算机:跨学科应用)3区
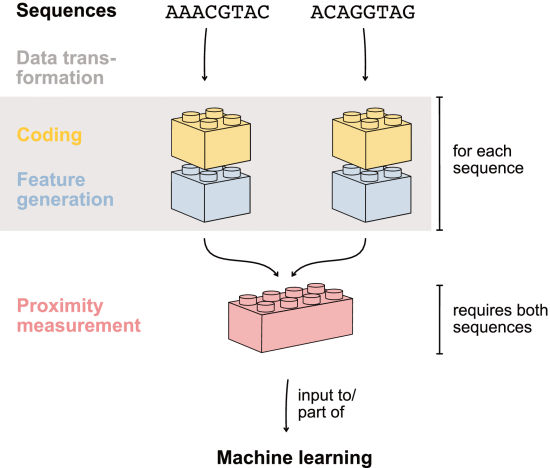
- 核心内容:序列映射到特征空间并在特征空间中进行比较:序列 --> 数值编码、特征生成 --> 问题特异性度量衡量相似性
-
作者信息:

-
标记蓝色说明是近5年发表在比较好的期刊上的论文
开篇:AF方法可用于:
alignment-free method出现:
[16] G. Kucherov, “Evolution of biosequence search algorithms: A brief survey,” Bioinformatics, vol. 35, no. 19, pp. 3547–3552, Oct. 2019.
AF方法推断系统发育:
[17] B. Haubold, “Alignment-free phylogenetics and population genetics,” Briefings Bioinf., vol. 15, no. 3, pp. 407–418, May 2014.
[18] C. X. Chan, G. Bernard, O. Poirion, J. M. Hogan, and M. A. Ragan, “Inferring phylogenies of evolving sequences without multiple sequence alignment,” Sci. Rep., vol. 4, no. 1, May 2015, Art. no. 6504.
[19] K. Hatje and M. Kollmar, “A phylogenetic analysis of the brassi- cales clade based on an alignment-free sequence comparison method,” Front. Plant Sci., vol. 3, 2012, Art. no. 192.
AF方法用于元基因组:
[20] Y.-W. Wu and Y. Ye, “A novel abundance-based algorithm for binning metagenomic sequences using l-tuples,” J. Comput. Biol.: A J. Comput. Mol. Cell Biol., vol. 18, no. 3, pp. 523–534, Mar. 2011.
[21] G. Leung and M. B. Eisen, “Identifying Cis-regulatory sequences by word profile similarity,” PLoS One, vol. 4, no. 9, Sep. 2009, Art. no. e6901.
AF方法用于数据库搜索:
[22] B. T. et al., “RAFTS3G: An efficient and versatile clustering soft- ware to analyses in large protein datasets,” BMC Bioinf., vol. 20, no. 1, Jul. 2019, Art. no. 392.
AF方法用于N








 最低0.47元/天 解锁文章
最低0.47元/天 解锁文章

















 1129
1129

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








