OpenCV机器学习库—支持向量机 SVM
文章目录
前言
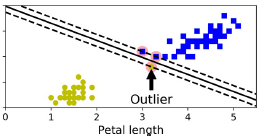
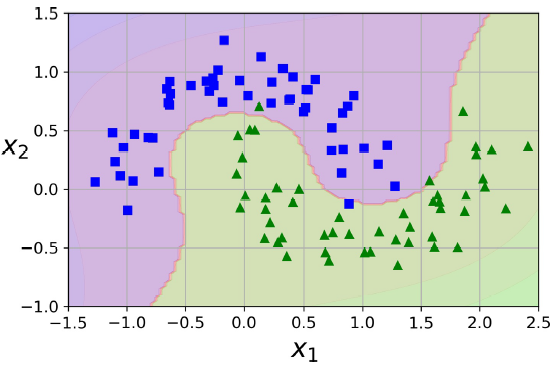
支持向量机主要引用于分类问题,而且主要针对二分类。另外SVM也可以扩展到多分类问题上,通过核函数的使用,将特定维度的数据映射到高维度的空间中。SVM分类器会找到一个分界面或者分界函数,使得两个类到这个分界面的距离和最大,所以也被称为“最大化类间距分类器”。可以通过设置核类型来适应特定维度的数据集
线性核:
非线性核:
一、支持向量机 SVM 定义
opencv中SVM的定义:opencv/ml.hpp
class CV_EXPORTS_W SVM : public StatModel
{
public:
class CV_EXPORTS Kernel : public Algorithm
{
public:
virtual int getType() const = 0;
virtual void calc( int vcount, int n, const float* vecs, const float* another, float* results ) = 0;
};
/** Type of a %SVM formulation.
See SVM::Types. Default value is SVM::C_SVC. */
/** @see setType */
CV_WRAP virtual int getType() const = 0;
/** @copybrief getType @see getType */
CV_WRAP virtual void setType(int val) = 0;
/** Parameter \f$\gamma\f$ of a kernel function.
For SVM::POLY, SVM::RBF, SVM::SIGMOID or SVM::CHI2. Default value is 1. */
/** @see setGamma */
CV_WRAP virtual double getGamma() const = 0;
/** @copybrief getGamma @see getGamma */
CV_WRAP virtual void setGamma(double val) = 0;
/** Parameter _coef0_ of a kernel function.
For SVM::POLY or SVM::SIGMOID. Default value is 0.*/
/** @see setCoef0 */
CV_WRAP virtual double getCoef0() const = 0;
/** @copybrief getCoef0 @see getCoef0 */
CV_WRAP virtual void setCoef0(double val) = 0;
/** Parameter _degree_ of a kernel function.
For SVM::POLY. Default value is 0. */
/** @see setDegree */
CV_WRAP virtual double getDegree() const = 0;
/** @copybrief getDegree @see getDegree */
CV_WRAP virtual void setDegree(double val) = 0;
/** Parameter _C_ of a %SVM optimization problem.
For SVM::C_SVC, SVM::EPS_SVR or SVM::NU_SVR. Default value is 0. */
/** @see setC */
CV_WRAP virtual double getC() const = 0;
/** @copybrief getC @see getC */
CV_WRAP virtual void setC(double val) = 0;
/** Parameter \f$\nu\f$ of a %SVM optimization problem.
For SVM::NU_SVC, SVM::ONE_CLASS or SVM::NU_SVR. Default value is 0. */
/** @see setNu */
CV_WRAP virtual double getNu() const = 0;
/** @copybrief getNu @see getNu */
CV_WRAP virtual void setNu(double val) = 0;
/** Parameter \f$\epsilon\f$ of a %SVM optimization problem.
For SVM::EPS_SVR. Default value is 0. */
/** @see setP */
CV_WRAP virtual double getP() const = 0;
/** @copybrief getP @see getP */
CV_WRAP virtual void setP(double val) = 0;
/** Optional weights in the SVM::C_SVC problem, assigned to particular classes.
They are multiplied by _C_ so the parameter _C_ of class _i_ becomes `classWeights(i) * C`. Thus
these weights affect the misclassification penalty for different classes. The larger weight,
the larger penalty on misclassification of data from the corresponding class. Default value is
empty Mat. */
/** @see setClassWeights */
CV_WRAP virtual cv::Mat getClassWeights() const = 0;
/** @copybrief getClassWeights @see getClassWeights */
CV_WRAP virtual void setClassWeights(const cv::Mat &val) = 0;
/** Termination criteria of the iterative %SVM training procedure which solves a partial
case of constrained quadratic optimization problem.
You can specify tolerance and/or the maximum number of iterations. Default value is
`TermCriteria( TermCriteria::MAX_ITER + TermCriteria::EPS, 1000, FLT_EPSILON )`; */
/** @see setTermCriteria */
CV_WRAP virtual cv::TermCriteria getTermCriteria() const = 0;
/** @copybrief getTermCriteria @see getTermCriteria */
CV_WRAP virtual void setTermCriteria(const cv::TermCriteria &val) = 0;
/** Type of a %SVM kernel.
See SVM::KernelTypes. Default value is SVM::RBF. */
CV_WRAP virtual int getKernelType() const = 0;
/** Initialize with one of predefined kernels.
See SVM::KernelTypes. */
CV_WRAP virtual void setKernel(int kernelType) = 0;
/** Initialize with custom kernel.
See SVM::Kernel class for implementation details */
virtual void setCustomKernel(const Ptr<Kernel> &_kernel) = 0;
//! %SVM type
enum Types {
/** C-Support Vector Classification. n-class classification (n \f$\geq\f$ 2), allows
imperfect separation of classes with penalty multiplier C for outliers. */
C_SVC=100,
/** \f$\nu\f$-Support Vector Classification. n-class classification with possible
imperfect separation. Parameter \f$\nu\f$ (in the range 0..1, the larger the value, the smoother
the decision boundary) is used instead of C. */
NU_SVC=101,
/** Distribution Estimation (One-class %SVM). All the training data are from
the same class, %SVM builds a boundary that separates the class from the rest of the feature
space. */
ONE_CLASS=102,
/** \f$\epsilon\f$-Support Vector Regression. The distance between feature vectors
from the training set and the fitting hyper-plane must be less than p. For outliers the
penalty multiplier C is used. */
EPS_SVR=103,
/** \f$\nu\f$-Support Vector Regression. \f$\nu\f$ is used instead of p.
See @cite LibSVM for details. */
NU_SVR=104
};
/** @brief %SVM kernel type
A comparison of different kernels on the following 2D test case with four classes. Four
SVM::C_SVC SVMs have been trained (one against rest) with auto_train. Evaluation on three
different kernels (SVM::CHI2, SVM::INTER, SVM::RBF). The color depicts the class with max score.
Bright means max-score \> 0, dark means max-score \< 0.

*/
enum KernelTypes {
/** Returned by SVM::getKernelType in case when custom kernel has been set */
CUSTOM=-1,
/** Linear kernel. No mapping is done, linear discrimination (or regression) is
done in the original feature space. It is the fastest option. \f$K(x_i, x_j) = x_i^T x_j\f$. */
LINEAR=0,
/** Polynomial kernel:
\f$K(x_i, x_j) = (\gamma x_i^T x_j + coef0)^{degree}, \gamma > 0\f$. */
POLY=1,
/** Radial basis function (RBF), a good choice in most cases.
\f$K(x_i, x_j) = e^{-\gamma ||x_i - x_j||^2}, \gamma > 0\f$. */
RBF=2,
/** Sigmoid kernel: \f$K(x_i, x_j) = \tanh(\gamma x_i^T x_j + coef0)\f$. */
SIGMOID=3,
/** Exponential Chi2 kernel, similar to the RBF kernel:
\f$K(x_i, x_j) = e^{-\gamma \chi^2(x_i,x_j)}, \chi^2(x_i,x_j) = (x_i-x_j)^2/(x_i+x_j), \gamma > 0\f$. */
CHI2=4,
/** Histogram intersection kernel. A fast kernel. \f$K(x_i, x_j) = min(x_i,x_j)\f$. */
INTER=5
};
//! %SVM params type
enum ParamTypes {
C=0,
GAMMA=1,
P=2,
NU=3,
COEF=4,
DEGREE=5
};
/** @brief Trains an %SVM with optimal parameters.
@param data the training data that can be constructed using TrainData::create or
TrainData::loadFromCSV.
@param kFold Cross-validation parameter. The training set is divided into kFold subsets. One
subset is used to test the model, the others form the train set. So, the %SVM algorithm is
executed kFold times.
@param Cgrid grid for C
@param gammaGrid grid for gamma
@param pGrid grid for p
@param nuGrid grid for nu
@param coeffGrid grid for coeff
@param degreeGrid grid for degree
@param balanced If true and the problem is 2-class classification then the method creates more
balanced cross-validation subsets that is proportions between classes in subsets are close
to such proportion in the whole train dataset.
The method trains the %SVM model automatically by choosing the optimal parameters C, gamma, p,
nu, coef0, degree. Parameters are considered optimal when the cross-validation
estimate of the test set error is minimal.
If there is no need to optimize a parameter, the corresponding grid step should be set to any
value less than or equal to 1. For example, to avoid optimization in gamma, set `gammaGrid.step
= 0`, `gammaGrid.minVal`, `gamma_grid.maxVal` as arbitrary numbers. In this case, the value
`Gamma` is taken for gamma.
And, finally, if the optimization in a parameter is required but the corresponding grid is
unknown, you may call the function SVM::getDefaultGrid. To generate a grid, for example, for
gamma, call `SVM::getDefaultGrid(SVM::GAMMA)`.
This function works for the classification (SVM::C_SVC or SVM::NU_SVC) as well as for the
regression (SVM::EPS_SVR or SVM::NU_SVR). If it is SVM::ONE_CLASS, no optimization is made and
the usual %SVM with parameters specified in params is executed.
*/
virtual bool trainAuto( const Ptr<TrainData>& data, int kFold = 10,
ParamGrid Cgrid = getDefaultGrid(C),
ParamGrid gammaGrid = getDefaultGrid(GAMMA),
ParamGrid pGrid = getDefaultGrid(P),
ParamGrid nuGrid = getDefaultGrid(NU),
ParamGrid coeffGrid = getDefaultGrid(COEF),
ParamGrid degreeGrid = getDefaultGrid(DEGREE),
bool balanced=false) = 0;
/** @brief Trains an %SVM with optimal parameters
@param samples training samples
@param layout See ml::SampleTypes.
@param responses vector of responses associated with the training samples.
@param kFold Cross-validation parameter. The training set is divided into kFold subsets. One
subset is used to test the model, the others form the train set. So, the %SVM algorithm is
@param Cgrid grid for C
@param gammaGrid grid for gamma
@param pGrid grid for p
@param nuGrid grid for nu
@param coeffGrid grid for coeff
@param degreeGrid grid for degree
@param balanced If true and the problem is 2-class classification then the method creates more
balanced cross-validation subsets that is proportions between classes in subsets are close
to such proportion in the whole train dataset.
The method trains the %SVM model automatically by choosing the optimal parameters C, gamma, p,
nu, coef0, degree. Parameters are considered optimal when the cross-validation
estimate of the test set error is minimal.
This function only makes use of SVM::getDefaultGrid for parameter optimization and thus only
offers rudimentary parameter options.
This function works for the classification (SVM::C_SVC or SVM::NU_SVC) as well as for the
regression (SVM::EPS_SVR or SVM::NU_SVR). If it is SVM::ONE_CLASS, no optimization is made and
the usual %SVM with parameters specified in params is executed.
*/
CV_WRAP virtual bool trainAuto(InputArray samples,
int layout,
InputArray responses,
int kFold = 10,
Ptr<ParamGrid> Cgrid = SVM::getDefaultGridPtr(SVM::C),
Ptr<ParamGrid> gammaGrid = SVM::getDefaultGridPtr(SVM::GAMMA),
Ptr<ParamGrid> pGrid = SVM::getDefaultGridPtr(SVM::P),
Ptr<ParamGrid> nuGrid = SVM::getDefaultGridPtr(SVM::NU),
Ptr<ParamGrid> coeffGrid = SVM::getDefaultGridPtr(SVM::COEF),
Ptr<ParamGrid> degreeGrid = SVM::getDefaultGridPtr(SVM::DEGREE),
bool balanced=false) = 0;
/** @brief Retrieves all the support vectors
The method returns all the support vectors as a floating-point matrix, where support vectors are
stored as matrix rows.
*/
CV_WRAP virtual Mat getSupportVectors() const = 0;
/** @brief Retrieves all the uncompressed support vectors of a linear %SVM
The method returns all the uncompressed support vectors of a linear %SVM that the compressed
support vector, used for prediction, was derived from. They are returned in a floating-point
matrix, where the support vectors are stored as matrix rows.
*/
CV_WRAP virtual Mat getUncompressedSupportVectors() const = 0;
/** @brief Retrieves the decision function
@param i the index of the decision function. If the problem solved is regression, 1-class or
2-class classification, then there will be just one decision function and the index should
always be 0. Otherwise, in the case of N-class classification, there will be \f$N(N-1)/2\f$
decision functions.
@param alpha the optional output vector for weights, corresponding to different support vectors.
In the case of linear %SVM all the alpha's will be 1's.
@param svidx the optional output vector of indices of support vectors within the matrix of
support vectors (which can be retrieved by SVM::getSupportVectors). In the case of linear
%SVM each decision function consists of a single "compressed" support vector.
The method returns rho parameter of the decision function, a scalar subtracted from the weighted
sum of kernel responses.
*/
CV_WRAP virtual double getDecisionFunction(int i, OutputArray alpha, OutputArray svidx) const = 0;
/** @brief Generates a grid for %SVM parameters.
@param param_id %SVM parameters IDs that must be one of the SVM::ParamTypes. The grid is
generated for the parameter with this ID.
The function generates a grid for the specified parameter of the %SVM algorithm. The grid may be
passed to the function SVM::trainAuto.
*/
static ParamGrid getDefaultGrid( int param_id );
/** @brief Generates a grid for %SVM parameters.
@param param_id %SVM parameters IDs that must be one of the SVM::ParamTypes. The grid is
generated for the parameter with this ID.
The function generates a grid pointer for the specified parameter of the %SVM algorithm.
The grid may be passed to the function SVM::trainAuto.
*/
CV_WRAP static Ptr<ParamGrid> getDefaultGridPtr( int param_id );
/** Creates empty model.
Use StatModel::train to train the model. Since %SVM has several parameters, you may want to
find the best parameters for your problem, it can be done with SVM::trainAuto. */
CV_WRAP static Ptr<SVM> create();
/** @brief Loads and creates a serialized svm from a file
*
* Use SVM::save to serialize and store an SVM to disk.
* Load the SVM from this file again, by calling this function with the path to the file.
*
* @param filepath path to serialized svm
*/
CV_WRAP static Ptr<SVM> load(const String& filepath);
};
opencv中SVM可用的核类型:
enum KernelTypes {
/** Returned by SVM::getKernelType in case when custom kernel has been set */
CUSTOM=-1,
/** Linear kernel. No mapping is done, linear discrimination (or regression) is
done in the original feature space. It is the fastest option. \f$K(x_i, x_j) = x_i^T x_j\f$. */
LINEAR=0,
/** Polynomial kernel:
\f$K(x_i, x_j) = (\gamma x_i^T x_j + coef0)^{degree}, \gamma > 0\f$. */
POLY=1,
/** Radial basis function (RBF), a good choice in most cases.
\f$K(x_i, x_j) = e^{-\gamma ||x_i - x_j||^2}, \gamma > 0\f$. */
RBF=2,
/** Sigmoid kernel: \f$K(x_i, x_j) = \tanh(\gamma x_i^T x_j + coef0)\f$. */
SIGMOID=3,
/** Exponential Chi2 kernel, similar to the RBF kernel:
\f$K(x_i, x_j) = e^{-\gamma \chi^2(x_i,x_j)}, \chi^2(x_i,x_j) = (x_i-x_j)^2/(x_i+x_j), \gamma > 0\f$. */
CHI2=4,
/** Histogram intersection kernel. A fast kernel. \f$K(x_i, x_j) = min(x_i,x_j)\f$. */
INTER=5
};
| 核 | OpenCV定义 |
|---|---|
| 线性 | cv::ml::SVM::LINEAR |
| 多项式 | cv::ml::SVM:: POLY |
| 径向基函数 | cv::ml::SVM:: RBF |
| Sigmoid | cv::ml::SVM::SIGMOID |
| 指数卡方分布 | cv::ml::SVM::GHI2 |
| 直方图交运算 | cv::ml::SVM:: INTER |
opencv中SVM可用的分类器类型:
enum Types {
/** C-Support Vector Classification. n-class classification (n \f$\geq\f$ 2), allows
imperfect separation of classes with penalty multiplier C for outliers. */
C_SVC=100,
/** \f$\nu\f$-Support Vector Classification. n-class classification with possible
imperfect separation. Parameter \f$\nu\f$ (in the range 0..1, the larger the value, the smoother
the decision boundary) is used instead of C. */
NU_SVC=101,
/** Distribution Estimation (One-class %SVM). All the training data are from
the same class, %SVM builds a boundary that separates the class from the rest of the feature
space. */
ONE_CLASS=102,
/** \f$\epsilon\f$-Support Vector Regression. The distance between feature vectors
from the training set and the fitting hyper-plane must be less than p. For outliers the
penalty multiplier C is used. */
EPS_SVR=103,
/** \f$\nu\f$-Support Vector Regression. \f$\nu\f$ is used instead of p.
See @cite LibSVM for details. */
NU_SVR=104
};
| SVM类型 | OpenCV中命名 |
|---|---|
| C-SVM分类器 | cv::ml::SVM::C_SVC |
| v-SVM分类器 | cv::ml::SVM:: NU_SVC |
| 一分类SVM | cv::ml::SVM:: ONE_CLASS |
| ε_SVR | cv::ml::SVM::EPS_SVR |
| v-SVR | cv::ml::SVM::NU_SVR |
解释:
1.c-SVM也叫“软边距SVM”,允许通过对离群点(不能被正确分类的数据点或者在核空间不能用超平面分割的数据集)的距离计算增加一定比例的处罚量来允许离群点的存在。
2.v-SVM相对于前者的固定的比例常量,引入了一个新的变量v,设定训练错误的上限,一般设在0-1之间。
3.多分类扩展SVM,OpenCV采用一种一对一的方法来实现多分类,当然还有一对多的方法。如果有K个类别,则有k(k-1)/2个分类器被训练,分类一个目标时,所有的分类器会一起工作
4.一分类SVM只有一个类,所有训练的数据都认为是一个类,其他不同的类会被区别开。
支持向量回归SVR,同样有两种ε_SVR和v-SVR,从本质上来说是一致的。
二、Opencv中SVM的使用
参数的设置
无论选择哪种SVM的类型,都需要相关的set函数来设置核函数
CV_WRAP virtual void setType(int val) ;
/** Parameter \f$\gamma\f$ of a kernel function.
For SVM::POLY, SVM::RBF, SVM::SIGMOID or SVM::CHI2. Default value is 1. */
CV_WRAP virtual void setGamma(double val) ;
/** Parameter _coef0_ of a kernel function.
For SVM::POLY or SVM::SIGMOID. Default value is 0.*/
CV_WRAP virtual void setCoef0(double val) ;
/** Parameter _degree_ of a kernel function.
For SVM::POLY. Default value is 0. */
CV_WRAP virtual void setDegree(double val) ;
/** Parameter _C_ of a %SVM optimization problem.
For SVM::C_SVC, SVM::EPS_SVR or SVM::NU_SVR. Default value is 0. */
CV_WRAP virtual void setC(double val) ;
/** Parameter \f$\nu\f$ of a %SVM optimization problem.
For SVM::NU_SVC, SVM::ONE_CLASS or SVM::NU_SVR. Default value is 0. */
CV_WRAP virtual void setNu(double val);
/** Parameter \f$\epsilon\f$ of a %SVM optimization problem.
For SVM::EPS_SVR. Default value is 0. */
CV_WRAP virtual void setP(double val) ;
CV_WRAP virtual void setClassWeights(const cv::Mat &val) ;
/** Termination criteria of the iterative %SVM training procedure which solves a partial
case of constrained quadratic optimization problem.
You can specify tolerance and/or the maximum number of iterations. Default value is
`TermCriteria( TermCriteria::MAX_ITER + TermCriteria::EPS, 1000, FLT_EPSILON )`; */
/** @see setTermCriteria */
CV_WRAP virtual void setTermCriteria(const cv::TermCriteria &val) ;
/** Initialize with one of predefined kernels.
See SVM::KernelTypes. */
CV_WRAP virtual void setKernel(int kernelType) ;
/** Initialize with custom kernel.
See SVM::Kernel class for implementation details */
virtual void setCustomKernel(const Ptr<Kernel> &_kernel) ;
Tips:
1.方法setClassWeights()允许用户通过提供一个单独的列向量来指定松弛参数的重要性,仅对C-SVM有效。提供的参数会与C相乘,这样用户可以对训练集中某些子集赋予更大的权重。默认不会使用。
2.方法setTermCriteria(),默认的参数为迭代1000次和FLT_EPSILON是安全的。
3.可以使用cv::ml::SVM::trainAuto()函数,它会尝试所有参数然后选择效果最好的参数,其用法和cv::ml::SVM::train()一样。
定义:
The method trains the %SVM model automatically by choosing the optimal parameters C, gamma, p,
nu, coef0, degree. Parameters are considered optimal when the cross-validation
estimate of the test set error is minimal.
This function only makes use of SVM::getDefaultGrid for parameter optimization and thus only
offers rudimentary parameter options.
This function works for the classification (SVM::C_SVC or SVM::NU_SVC) as well as for the
regression (SVM::EPS_SVR or SVM::NU_SVR). If it is SVM::ONE_CLASS, no optimization is made and
the usual %SVM with parameters specified in params is executed.
*/
CV_WRAP virtual bool trainAuto(InputArray samples,
int layout,
InputArray responses,
int kFold = 10,
Ptr<ParamGrid> Cgrid = SVM::getDefaultGridPtr(SVM::C),
Ptr<ParamGrid> gammaGrid = SVM::getDefaultGridPtr(SVM::GAMMA),
Ptr<ParamGrid> pGrid = SVM::getDefaultGridPtr(SVM::P),
Ptr<ParamGrid> nuGrid = SVM::getDefaultGridPtr(SVM::NU),
Ptr<ParamGrid> coeffGrid = SVM::getDefaultGridPtr(SVM::COEF),
Ptr<ParamGrid> degreeGrid = SVM::getDefaultGridPtr(SVM::DEGREE),
bool balanced=false) = 0;
预测
利用cv::ml::SVM::predict()函数预测:
float cv::ml::SVM::predict(
InputArray samples;
OutputArray results = noArray(),
int flags = 0
) const;
其他方法
/*The method returns all the support vectors as a floating-point matrix, where support vectors are
stored as matrix rows.*/
CV_WRAP virtual Mat getSupportVectors() const = 0;
/**The method returns all the uncompressed support vectors of a linear %SVM that the compressed
support vector, used for prediction, was derived from. They are returned in a floating-point
matrix, where the support vectors are stored as matrix rows.
*/
CV_WRAP virtual Mat getUncompressedSupportVectors() const = 0;
/*The method returns rho parameter of the decision function, a scalar subtracted from the weighted
sum of kernel responses.
*/
CV_WRAP virtual double getDecisionFunction(int i, OutputArray alpha, OutputArray svidx) const = 0;
getSupportVectors()函数返回所有用于决定超平面的支撑向量;
getUncompressedSupportVectors()函数返回原始的支撑向量;
getDecisionFunction()函数返回每一个决策平面或者超平面。
另外cv::ml::SVM提供了save()、load()、clear()函数来进行保存模型、加载模型和清除模型。
三、创建示例
//一、创建一个分类器
cv::Ptr<cv::ml::SVM> svm = cv::ml::SVM::create();
//二、设置相关参数
//设置类型
svm->setType(cv::ml::SVM::C_SVC);
//设置核类型
svm->setKernel(cv::ml::SVM::LINEAR);
//设置参数
svm->setGamma(1.0);
svm->setDegree(1.0);
svm->setCoef0(1);
svm->setNu(0.5);
svm->setP(0);
svm->setC(C);
//设置迭代条件
svm->setTermCriteria(cv::TermCriteria(cv::TermCriteria::MAX_ITER + cv::TermCriteria::EPS, 10000, 0.0001));
//三、数据集获取
cv::Ptr<cv::ml::TrainData> train_data = cv::ml::TrainData::create(datas,cv::ml::ROW_SAMPLE, labels)
//四、训练
svm->train(train_data);
//五、预测
float ret = svm->predict(test_images, pre_out);
//六、保存模型
svm->save("***.xml");
//七、加载模型
cv::Ptr<cv::ml::SVM> svm = cv::ml::SVM::load<cv::ml::SVM>("***.xml");
OpenCV例程1
位置::\samples\cpp\tutorial_code\ml\introduction_to_svm.cpp
#include <opencv2/core.hpp>
#include <opencv2/imgproc.hpp>
#include <opencv2/imgcodecs.hpp>
#include <opencv2/highgui.hpp>
#include <opencv2/ml.hpp>
using namespace cv;
using namespace cv::ml;
int main(int, char**)
{
// Set up training data
//! [setup1]
int labels[4] = {1, -1, -1, -1};
float trainingData[4][2] = { {501, 10}, {255, 10}, {501, 255}, {10, 501} };
//! [setup1]
//! [setup2]
Mat trainingDataMat(4, 2, CV_32F, trainingData);
Mat labelsMat(4, 1, CV_32SC1, labels);
//! [setup2]
// Train the SVM
//! [init]
Ptr<SVM> svm = SVM::create();
svm->setType(SVM::C_SVC);
svm->setKernel(SVM::LINEAR);
svm->setTermCriteria(TermCriteria(TermCriteria::MAX_ITER, 100, 1e-6));
//! [init]
//! [train]
svm->train(trainingDataMat, ROW_SAMPLE, labelsMat);
//! [train]
// Data for visual representation
int width = 512, height = 512;
Mat image = Mat::zeros(height, width, CV_8UC3);
// Show the decision regions given by the SVM
//! [show]
Vec3b green(0,255,0), blue(255,0,0);
for (int i = 0; i < image.rows; i++)
{
for (int j = 0; j < image.cols; j++)
{
Mat sampleMat = (Mat_<float>(1,2) << j,i);
float response = svm->predict(sampleMat);
if (response == 1)
image.at<Vec3b>(i,j) = green;
else if (response == -1)
image.at<Vec3b>(i,j) = blue;
}
}
//! [show]
// Show the training data
//! [show_data]
int thickness = -1;
circle( image, Point(501, 10), 5, Scalar( 0, 0, 0), thickness );
circle( image, Point(255, 10), 5, Scalar(255, 255, 255), thickness );
circle( image, Point(501, 255), 5, Scalar(255, 255, 255), thickness );
circle( image, Point( 10, 501), 5, Scalar(255, 255, 255), thickness );
//! [show_data]
// Show support vectors
//! [show_vectors]
thickness = 2;
Mat sv = svm->getUncompressedSupportVectors();
for (int i = 0; i < sv.rows; i++)
{
const float* v = sv.ptr<float>(i);
circle(image, Point( (int) v[0], (int) v[1]), 6, Scalar(128, 128, 128), thickness);
}
//! [show_vectors]
imwrite("result.png", image); // save the image
imshow("SVM Simple Example", image); // show it to the user
waitKey();
return 0;
}
OpenCV例程2
位置::samples\cpp\tutorial_code\ml\non_linear_svms.cpp
#include <iostream>
#include <opencv2/core.hpp>
#include <opencv2/imgproc.hpp>
#include "opencv2/imgcodecs.hpp"
#include <opencv2/highgui.hpp>
#include <opencv2/ml.hpp>
using namespace cv;
using namespace cv::ml;
using namespace std;
static void help()
{
cout<< "\n--------------------------------------------------------------------------" << endl
<< "This program shows Support Vector Machines for Non-Linearly Separable Data. " << endl
<< "--------------------------------------------------------------------------" << endl
<< endl;
}
int main()
{
help();
const int NTRAINING_SAMPLES = 100; // Number of training samples per class
const float FRAC_LINEAR_SEP = 0.9f; // Fraction of samples which compose the linear separable part
// Data for visual representation
const int WIDTH = 512, HEIGHT = 512;
Mat I = Mat::zeros(HEIGHT, WIDTH, CV_8UC3);
//--------------------- 1. Set up training data randomly ---------------------------------------
Mat trainData(2*NTRAINING_SAMPLES, 2, CV_32F);
Mat labels (2*NTRAINING_SAMPLES, 1, CV_32S);
RNG rng(100); // Random value generation class
// Set up the linearly separable part of the training data
int nLinearSamples = (int) (FRAC_LINEAR_SEP * NTRAINING_SAMPLES);
//! [setup1]
// Generate random points for the class 1
Mat trainClass = trainData.rowRange(0, nLinearSamples);
// The x coordinate of the points is in [0, 0.4)
Mat c = trainClass.colRange(0, 1);
rng.fill(c, RNG::UNIFORM, Scalar(0), Scalar(0.4 * WIDTH));
// The y coordinate of the points is in [0, 1)
c = trainClass.colRange(1,2);
rng.fill(c, RNG::UNIFORM, Scalar(0), Scalar(HEIGHT));
// Generate random points for the class 2
trainClass = trainData.rowRange(2*NTRAINING_SAMPLES-nLinearSamples, 2*NTRAINING_SAMPLES);
// The x coordinate of the points is in [0.6, 1]
c = trainClass.colRange(0 , 1);
rng.fill(c, RNG::UNIFORM, Scalar(0.6*WIDTH), Scalar(WIDTH));
// The y coordinate of the points is in [0, 1)
c = trainClass.colRange(1,2);
rng.fill(c, RNG::UNIFORM, Scalar(0), Scalar(HEIGHT));
//! [setup1]
//------------------ Set up the non-linearly separable part of the training data ---------------
//! [setup2]
// Generate random points for the classes 1 and 2
trainClass = trainData.rowRange(nLinearSamples, 2*NTRAINING_SAMPLES-nLinearSamples);
// The x coordinate of the points is in [0.4, 0.6)
c = trainClass.colRange(0,1);
rng.fill(c, RNG::UNIFORM, Scalar(0.4*WIDTH), Scalar(0.6*WIDTH));
// The y coordinate of the points is in [0, 1)
c = trainClass.colRange(1,2);
rng.fill(c, RNG::UNIFORM, Scalar(0), Scalar(HEIGHT));
//! [setup2]
//------------------------- Set up the labels for the classes ---------------------------------
labels.rowRange( 0, NTRAINING_SAMPLES).setTo(1); // Class 1
labels.rowRange(NTRAINING_SAMPLES, 2*NTRAINING_SAMPLES).setTo(2); // Class 2
//------------------------ 2. Set up the support vector machines parameters --------------------
cout << "Starting training process" << endl;
//! [init]
Ptr<SVM> svm = SVM::create();
svm->setType(SVM::C_SVC);
svm->setC(0.1);
svm->setKernel(SVM::LINEAR);
svm->setTermCriteria(TermCriteria(TermCriteria::MAX_ITER, (int)1e7, 1e-6));
//! [init]
//------------------------ 3. Train the svm ----------------------------------------------------
//! [train]
svm->train(trainData, ROW_SAMPLE, labels);
//! [train]
cout << "Finished training process" << endl;
//------------------------ 4. Show the decision regions ----------------------------------------
//! [show]
Vec3b green(0,100,0), blue(100,0,0);
for (int i = 0; i < I.rows; i++)
{
for (int j = 0; j < I.cols; j++)
{
Mat sampleMat = (Mat_<float>(1,2) << j, i);
float response = svm->predict(sampleMat);
if (response == 1) I.at<Vec3b>(i,j) = green;
else if (response == 2) I.at<Vec3b>(i,j) = blue;
}
}
//! [show]
//----------------------- 5. Show the training data --------------------------------------------
//! [show_data]
int thick = -1;
float px, py;
// Class 1
for (int i = 0; i < NTRAINING_SAMPLES; i++)
{
px = trainData.at<float>(i,0);
py = trainData.at<float>(i,1);
circle(I, Point( (int) px, (int) py ), 3, Scalar(0, 255, 0), thick);
}
// Class 2
for (int i = NTRAINING_SAMPLES; i <2*NTRAINING_SAMPLES; i++)
{
px = trainData.at<float>(i,0);
py = trainData.at<float>(i,1);
circle(I, Point( (int) px, (int) py ), 3, Scalar(255, 0, 0), thick);
}
//! [show_data]
//------------------------- 6. Show support vectors --------------------------------------------
//! [show_vectors]
thick = 2;
Mat sv = svm->getUncompressedSupportVectors();
for (int i = 0; i < sv.rows; i++)
{
const float* v = sv.ptr<float>(i);
circle(I, Point( (int) v[0], (int) v[1]), 6, Scalar(128, 128, 128), thick);
}
//! [show_vectors]
imwrite("result.png", I); // save the Image
imshow("SVM for Non-Linear Training Data", I); // show it to the user
waitKey();
return 0;
}






















 4699
4699











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








