1.安装
conda create -n cellDancer python==3.7.6
conda activate cellDancer
pip install celldancer2.准备数据,跟scVelo处理数据步骤相同,详见:单细胞 | RNA速率 · scVelo_rna速率 -velocyto-CSDN博客
# 将 adata 格式传输到 dataframe
import packages
import os
import sys
import glob
import pandas as pd
import math
import matplotlib.pyplot as plt
import celldancer as cd
import celldancer.cdplt as cdplt
from celldancer.cdplt import colormap
adata = scv.read('./celltypesc.h5ad')
cdutil.adata_to_df_with_embed(adata,
us_para=['Mu','Ms'],
cell_type_para='celltype',
embed_para='X_umap',
save_path='cell_type_u_s.csv')
cell_type_u_s_path = './cell_type_u_s.csv'
cell_type_u_s = pd.read_csv(cell_type_u_s_path)
cell_type_u_scellDancer 的输入数据包含‘gene_name’, ‘unsplice’, ‘splice’ ,‘cellID’ ,‘clusters’ ,‘embedding1’, and ‘embedding2.’。
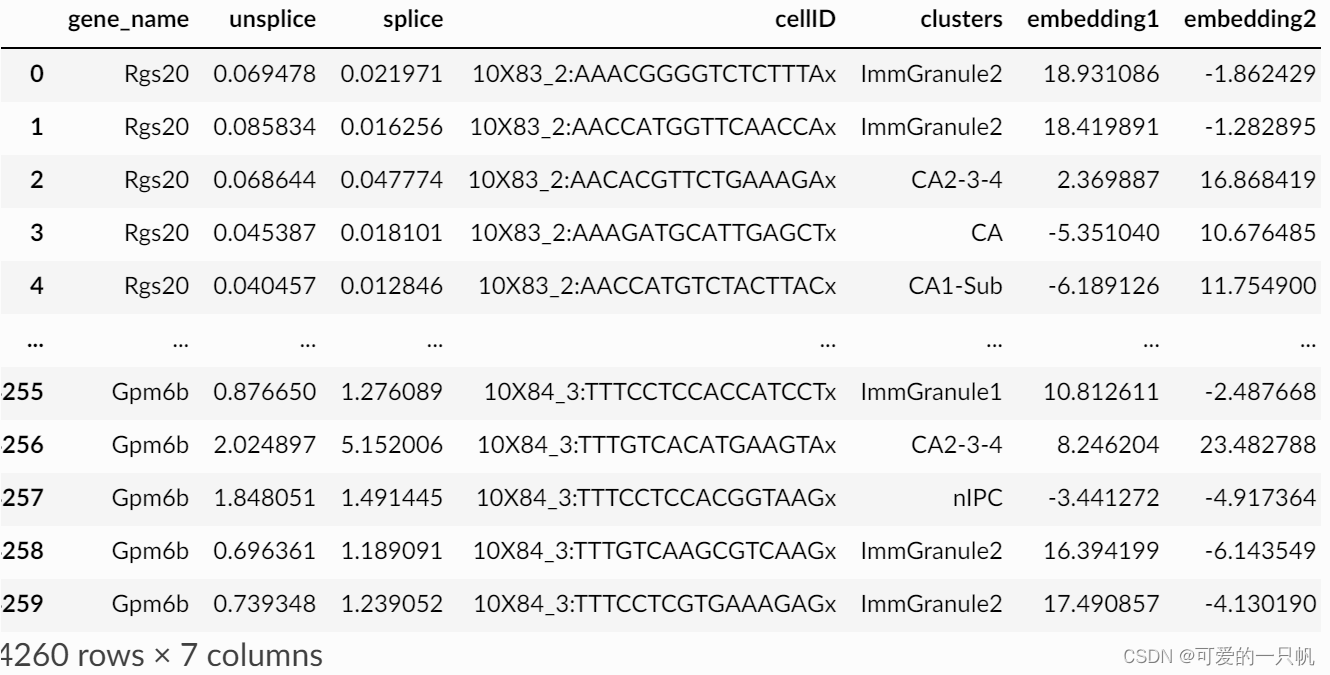
3.估计样本的RNA速率:与scVelo不同的是,cellDancer是对每个细胞的每个基因进行速率分析,可以指定你需要分析的genelist。
gene_list=['Psd3', 'Dcx', 'Syt11', 'Ntrk2', 'Gnao1',
'Gria1', 'Dctn3', 'Map1b', 'Camk2a', 'Gpm6b',
'Sez6l', 'Evl', 'Astn1', 'Ank2', 'Klf7',
'Tbc1d16', 'Atp1a3', 'Stxbp6', 'Scn2a1',
'Lhx9', 'Slc4a4', 'Ppfia2', 'Kcnip1', 'Ptpro',
'Diaph3', 'Slc1a3', 'Cadm1', 'Mef2c', 'Sptbn1', 'Ncald']
loss_df, cellDancer_df=cd.velocity(cell_type_u_s,
gene_list=gene_list,
permutation_ratio=0.1,
norm_u_s=False,
norm_cell_distribution=False,
n_jobs=8)
cellDancer_df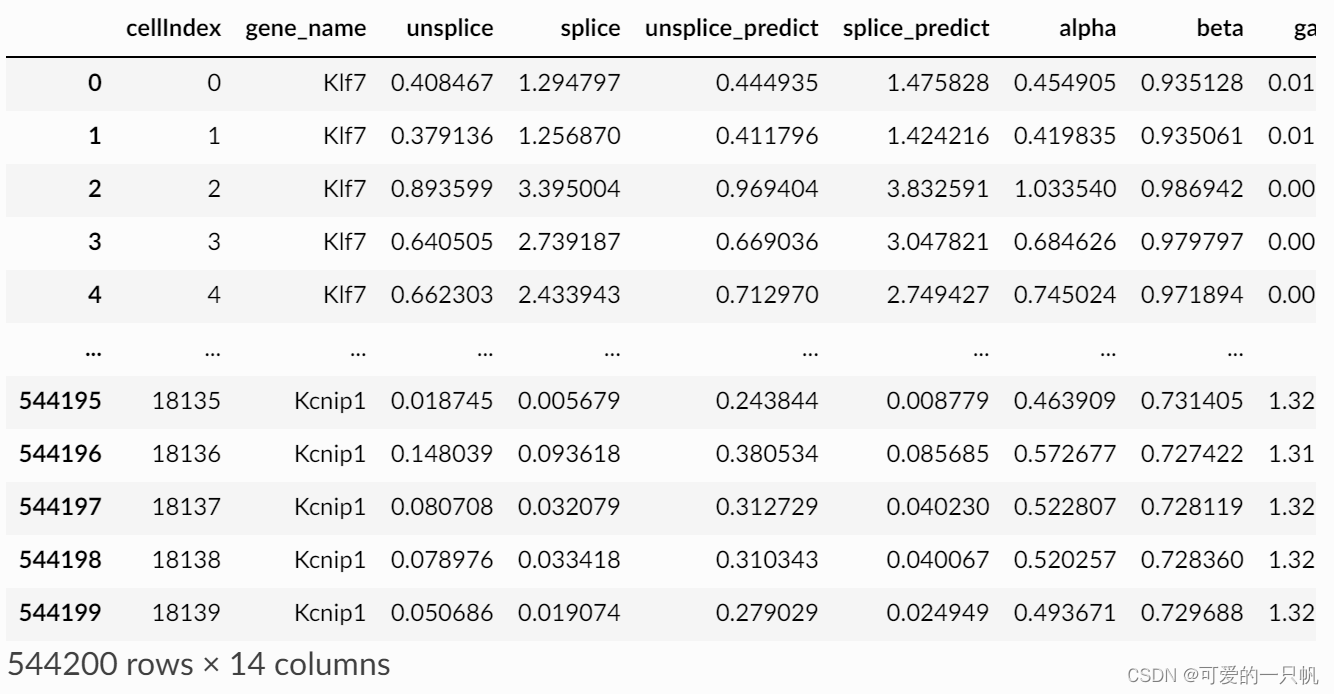
4.可视化1:RNA速率
# compute cell velocity
cellDancer_df=cd.compute_cell_velocity(cellDancer_df=cellDancer_df,
projection_neighbor_choice='gene',
expression_scale='power10',
projection_neighbor_size=10, speed_up=(100,100))
colevels=adata.obs.celltype.unique()
cellcolor=dict(zip(colevels,sns.color_palette("husl", len(colevels)).as_hex()))
# plot cell velocity
fig, ax = plt.subplots(figsize=(17,17))
cdplt.scatter_cell(ax,
cellDancer_df,
colors=cellcolor,
alpha=1,
s=10,
velocity=True,
legend='on',
min_mass=15,
arrow_grid=(30,30),
custom_xlim=[-13,13],
custom_ylim=[-16,16],
)
ax.axis('off')
plt.show()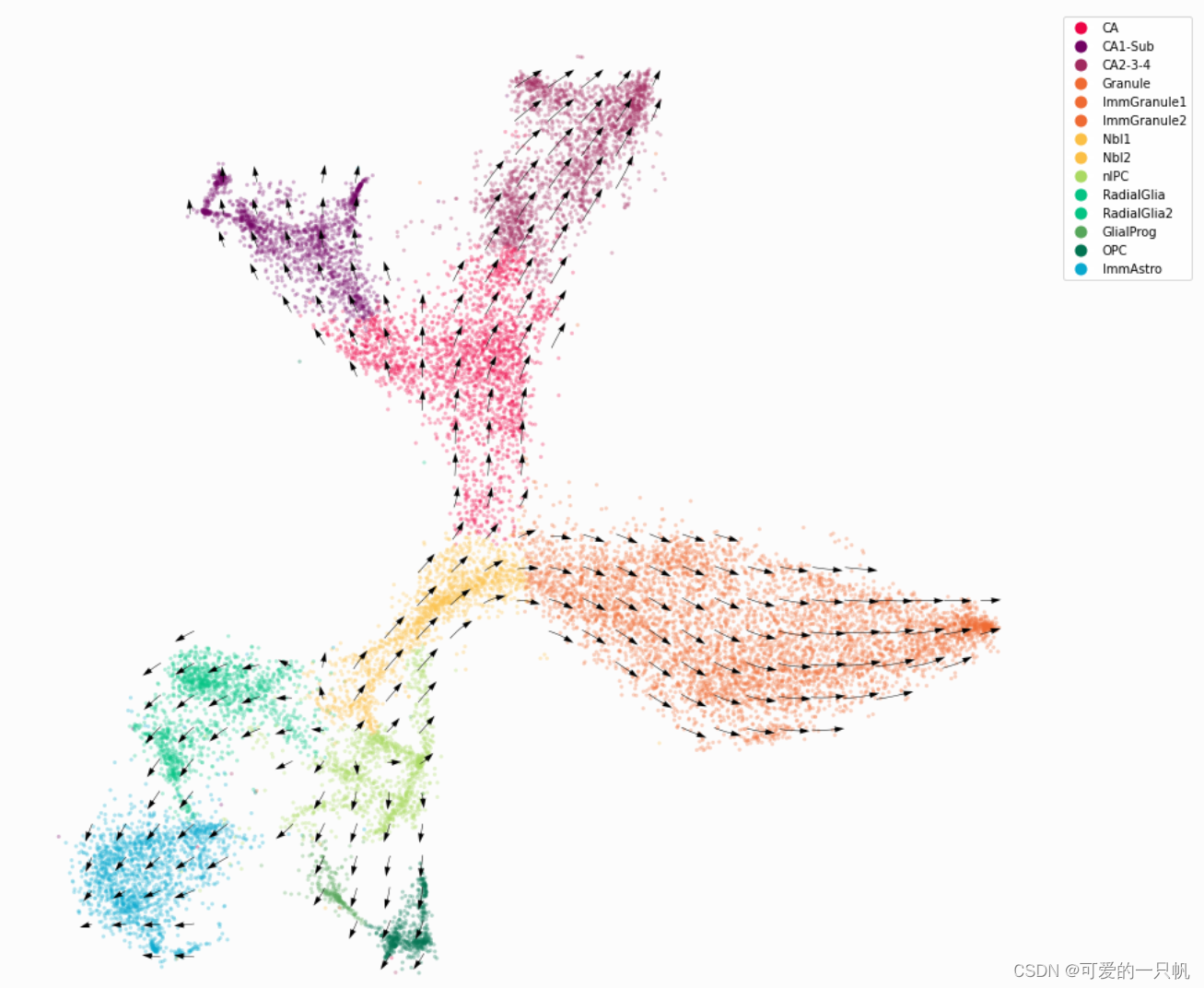
可视化2:每个基因的速率分析图
ncols=5
height=math.ceil(len(gene_list)/ncols)*4
fig = plt.figure(figsize=(20,height))
for i in range(len(gene_list)):
ax = fig.add_subplot(math.ceil(len(gene_list)/ncols), ncols, i+1)
cdplt.scatter_gene(
ax=ax,
x='splice',
y='unsplice',
cellDancer_df=cellDancer_df,
custom_xlim=None,
custom_ylim=None,
colors=colormap.colormap_neuro,
alpha=0.5,
s = 10,
velocity=True,
gene=gene_list[i])
ax.set_title(gene_list[i])
ax.axis('off')
plt.show()
可视化3:伪时序图
import random
# set parameters
dt = 0.05
t_total = {dt:int(10/dt)}
n_repeats = 10
# estimate pseudotime
cellDancer_df = cd.pseudo_time(cellDancer_df=cellDancer_df,
grid=(30,30),
dt=dt,
t_total=t_total[dt],
n_repeats=n_repeats,
speed_up=(100,100),
n_paths = 3,
plot_long_trajs=True,
psrng_seeds_diffusion=[i for i in range(n_repeats)],
n_jobs=8)
fig, ax = plt.subplots(figsize=(10,10))
im=cdplt.scatter_cell(ax,cellDancer_df, colors='pseudotime',
alpha=0.5, velocity=False)
ax.axis('off')
plt.show()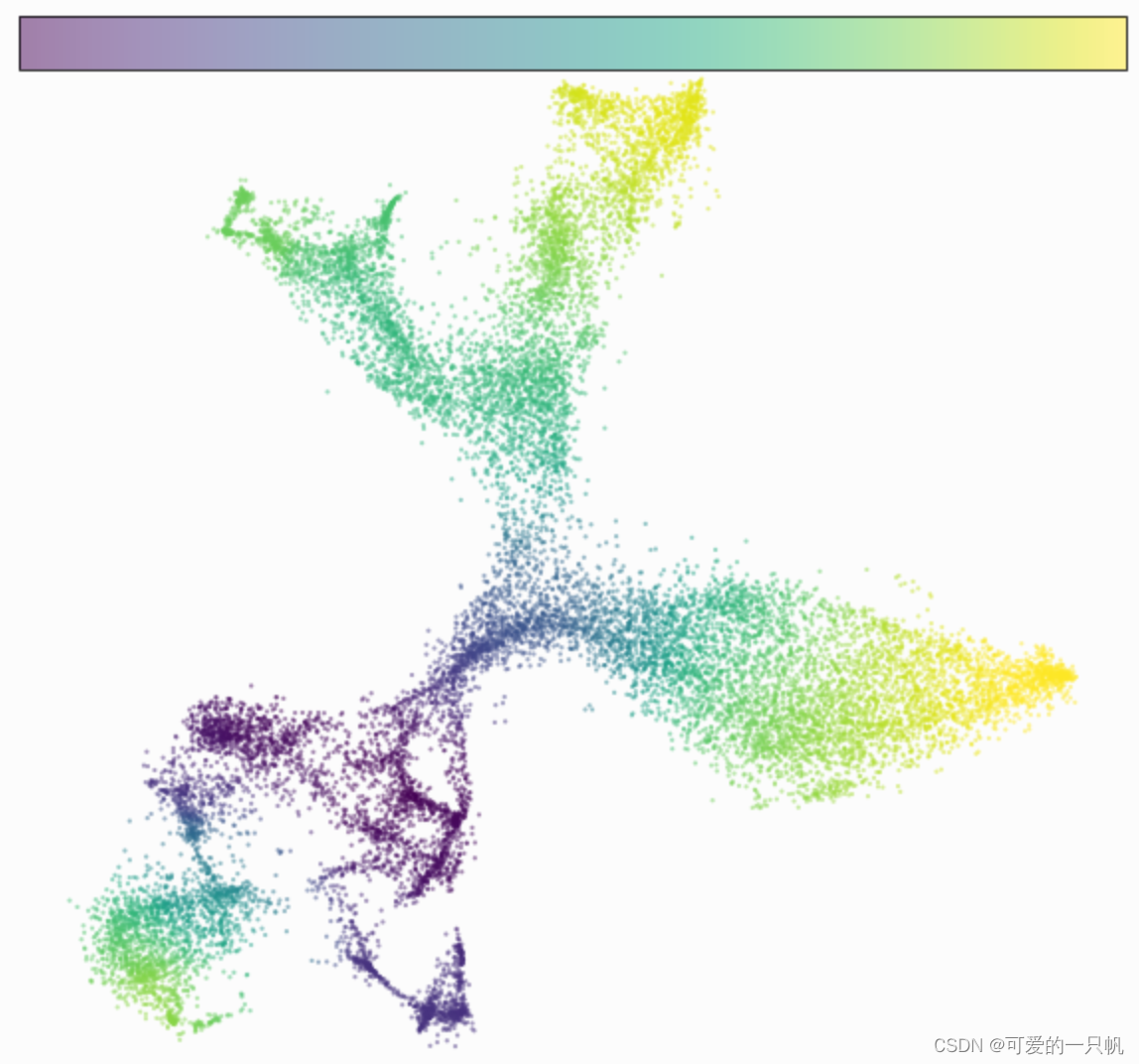
可视化4:显示剪接 RNA 沿伪时间的丰度
gene_list=['Psd3', 'Gria1', 'Camk2a', 'Sez6l', 'Ank2',
'Atp1a3', 'Stxbp6', 'Scn2a1', 'Ppfia2', 'Kcnip1',
'Ntrk2', 'Mef2c', 'Sptbn1', 'Ncald','Dcx',
'Syt11','Slc1a3', 'Dctn3', 'Map1b', 'Gpm6b',
'Evl', 'Astn1', 'Tbc1d16','Slc4a4', 'Ptpro']
ncols=5
height=math.ceil(len(gene_list)/ncols)*4
fig = plt.figure(figsize=(20,height))
for i in range(len(gene_list)):
ax = fig.add_subplot(math.ceil(len(gene_list)/ncols), ncols, i+1)
cdplt.scatter_gene(
ax=ax,
x='pseudotime',
y='splice',
cellDancer_df=cellDancer_df,
custom_xlim=None,
custom_ylim=None,
colors=colormap.colormap_neuro,
alpha=0.5,
s = 5,
velocity=False,
gene=gene_list[i])
ax.set_title(gene_list[i])
ax.axis('off')
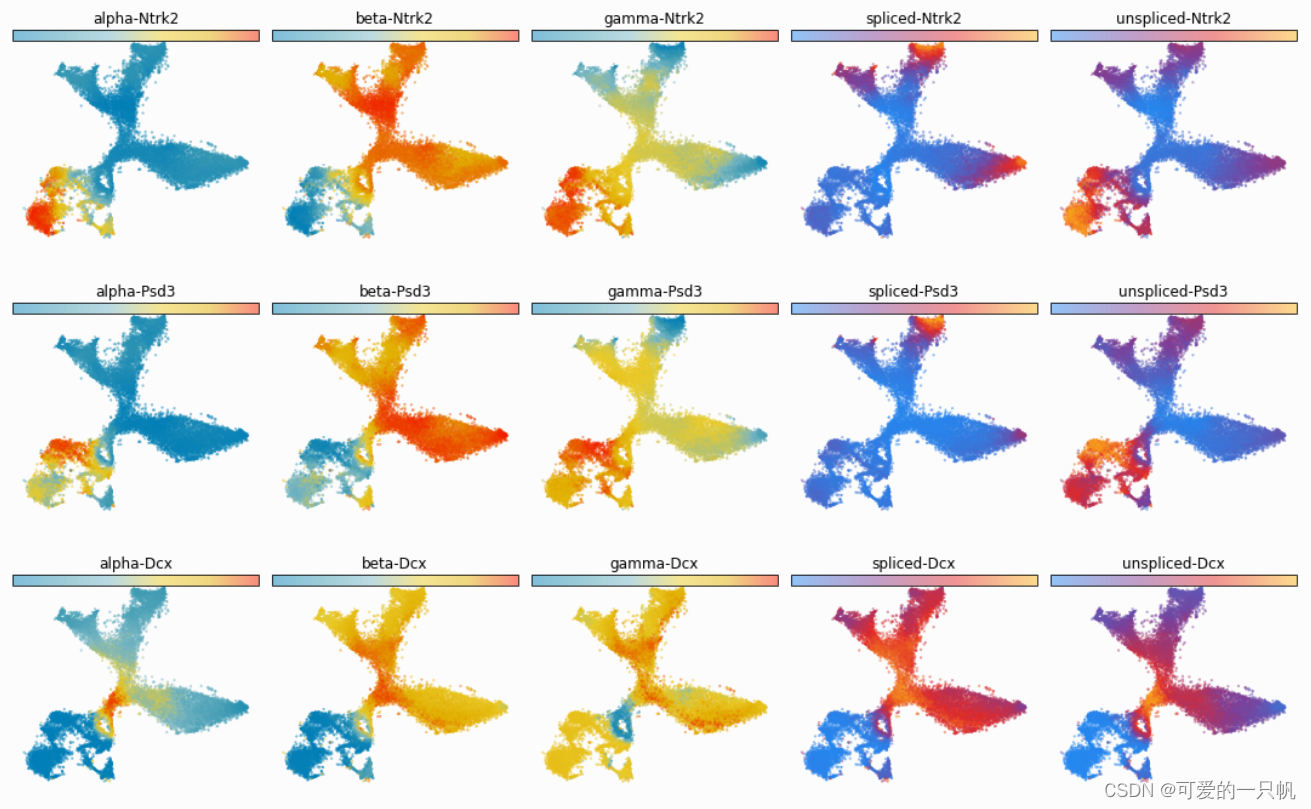
可视化5:展示预测动力学速率(转录α、剪接β和降解速率γ)、剪接 mRNA 丰度和未剪接 mRNA 丰度。
gene_samples=['Ntrk2','Psd3','Dcx']
for gene in gene_samples:
fig, ax = plt.subplots(ncols=5, figsize=(15,3))
cdplt.scatter_cell(ax[0],cellDancer_df, colors='alpha',
gene=gene, velocity=False)
cdplt.scatter_cell(ax[1],cellDancer_df, colors='beta',
gene=gene, velocity=False)
cdplt.scatter_cell(ax[2],cellDancer_df, colors='gamma',
gene=gene, velocity=False)
cdplt.scatter_cell(ax[3],cellDancer_df, colors='splice',
gene=gene, velocity=False)
cdplt.scatter_cell(ax[4],cellDancer_df, colors='unsplice',
gene=gene, velocity=False)
ax[0].axis('off')
ax[1].axis('off')
ax[2].axis('off')
ax[3].axis('off')
ax[4].axis('off')
ax[0].set_title('alpha-'+gene)
ax[1].set_title('beta-'+gene)
ax[2].set_title('gamma-'+gene)
ax[3].set_title('spliced-'+gene)
ax[4].set_title('unspliced-'+gene)
plt.tight_layout()
plt.show()





















 647
647











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








