一、环境准备及背景介绍
Python 开发环境:搭建 Python 高效开发环境: Pycharm + Anaconda
Biopython 序列处理:生物信息中的 Python 02 | 用biopython解析序列
示例 Genbank 数据:下载链接
Genbank 数据介绍:生物信息中的Python 05 | 从 Genbank 文件中提取 CDS 等其他特征序列
目录结构:

二、Python 实现
BaimoTools.py
#!/usr/bin/env python
# -*- coding: utf-8 -*-
# @Author : Baimoc
# @Email : baimoc@163.com
# @Time : 2020/12/20 14:28
# @File : BaimoTools
import os
import time
from Bio import SeqIO, SeqFeature
class BaimoTools():
def __init__(self, gb_file, fasta_file):
self.complete_fasta = ""
self.fasta_file = fasta_file
self.gb_file = gb_file
self.feature = None
self.record = None
def format_val(self, object=None):
"""
格式化对象值为字符串
:param object: 对象或对象键值
:return:
"""
key = ""
# 判断参数是否为字符
if isinstance(object, str):
obj = self.feature.qualifiers
key = object
else:
obj = object
# 为字符,提取 feature.qualifiers 对象关键字
if key != "" and not obj.get(key):
return ""
elif key == "":
val = obj
else:
val = obj[key]
# 转换为字符串
if not len(val):
val = ""
elif len(val) == 1:
val = val[0]
else:
if isinstance(val, SeqFeature.CompoundLocation) or isinstance(val, SeqFeature.FeatureLocation):
val = str(val)
else:
val = " | ".join(val)
return val
def extract_cds(self, cds):
"""
获取 CDS 的 Fasta 序列
:param cds: 获取指定基因的 CDS 区域,如果为空,则获取全部
"""
records = list(SeqIO.parse(self.gb_file, "genbank"))
for record in records:
print(f"{record.id}")
for feature in record.features:
# 提取 CDS 信息
if feature.type == "CDS":
self.feature = feature
self.record = record
cds_gene = self.format_val('gene')
if cds == "":
self.complete_fasta += self.format_fasta()
elif isinstance(cds, str) and cds_gene == cds:
self.complete_fasta += self.format_fasta()
elif isinstance(cds, list) and cds_gene in cds:
self.complete_fasta += self.format_fasta()
self.write_file()
def write_file(self):
"""
写入文件
"""
with open(self.fasta_file, "w") as f_obj:
f_obj.writelines(self.complete_fasta)
def format_fasta(self, num=0):
"""
整理 Fasta 格式
:param num: 每行字符数,超出则换行
:return: Fasta 文本
"""
cds_gene = self.format_val('gene')
cds_location = self.format_val(self.feature.location)
cds_product = self.format_val('product')
cds_protein_id = self.format_val('protein_id')
cds_translation = self.format_val('translation')
complete_ana = f">{self.record.id} | {cds_gene} | {cds_product} | {cds_protein_id} | {str(cds_location)}\n"
format_seq = ""
if num:
for i, char in enumerate(cds_translation):
format_seq += char
if (i + 1) % num == 0:
format_seq += "\n"
else:
format_seq = cds_translation
return complete_ana + format_seq + "\n"
三、使用示例
数据介绍
示例数据为新冠病毒的基因组 genbank 文件,文件中包含:
两个基因组:LC553263.1 和 LC553262.1
一个基因组会有多个基因,下面是它的基因组结构:
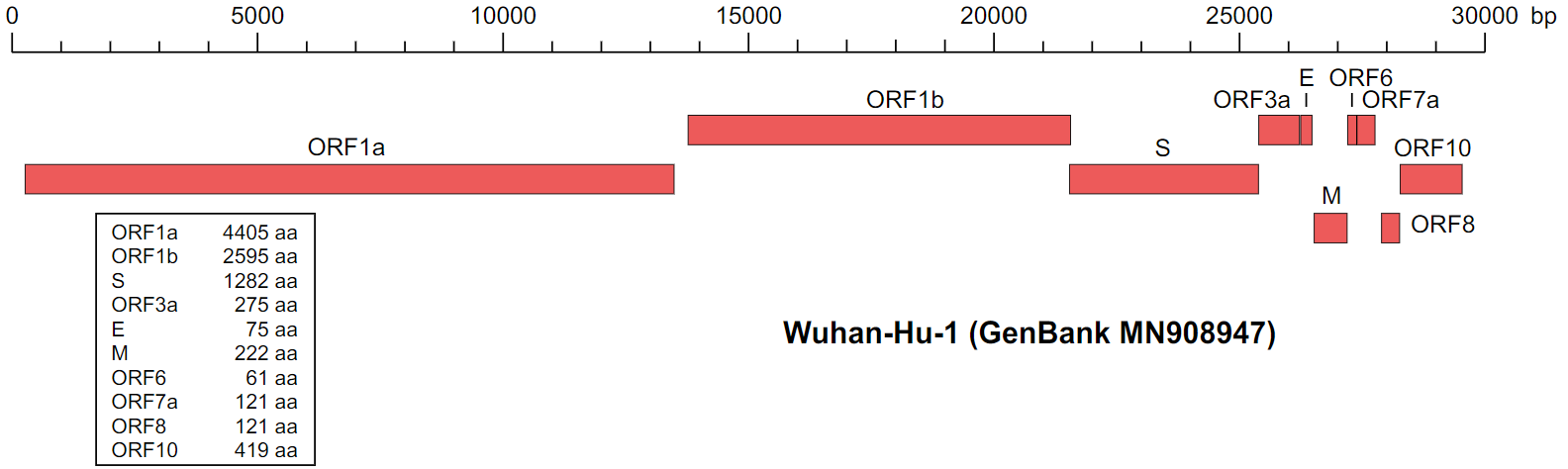
1、提取单个基因CDS
main.py
from BaimoTools import BaimoTools
gb_file = f"res/genbank/SARS-CoV-2.gb"
fasta_file = f"out/output_s.fasta"
baimoTools = BaimoTools(gb_file, fasta_file)
# baimoTools.extract_cds('S')
输出文件 output_s.fasta,分别提取到两个基因组的 S 基因 CDS 区域:

2、提取多个基因CDS
main.py
from BaimoTools import BaimoTools
gb_file = f"res/genbank/SARS-CoV-2.gb"
fasta_file = f"out/output_s_m_orf10.fasta"
baimoTools = BaimoTools(gb_file, fasta_file)
baimoTools.extract_cds(['S', 'M', 'ORF10'])
输出文件 output_s_m_orf10.fasta,分别提取到两个基因组的 S,M,ORF10 基因 CDS 区域::

2、提取全部基因CDS
main.py
from BaimoTools import BaimoTools
gb_file = f"res/genbank/SARS-CoV-2.gb"
fasta_file = f"out/output_s.fasta"
baimoTools = BaimoTools(gb_file, fasta_file)
# baimoTools.extract_cds("")
输出文件 output_all.fasta,分别提取到两个基因组的全部基因 CDS 区域:

下一步更新其他基因特征提取,及格式转换功能
我的博客即将同步至腾讯云+社区,邀请大家一同入驻:https://cloud.tencent.com/developer/support-plan?invite_code=1cy01nvbylvi6



























 3767
3767

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










