一边学习,一边总结,一边分享!
写在前面
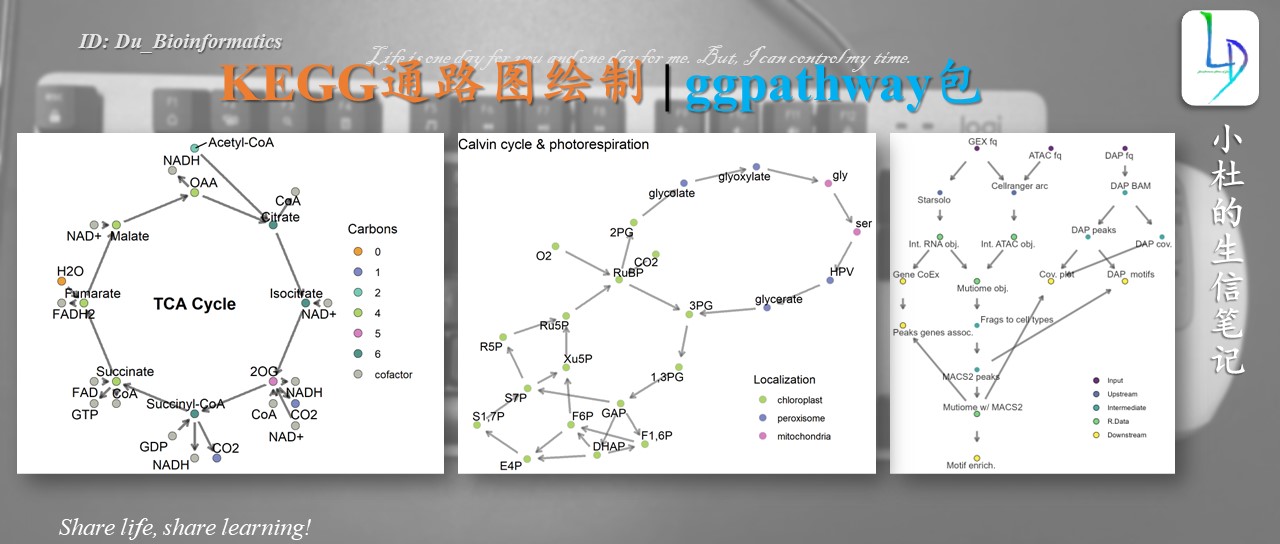
今天在GitHub中看到一个ggpathway的包,主要可以制作通路网络图,或是进一步优化的话,可以进行个性话制作。
操作步骤在GitHub中已经很详细。自己也照葫芦画瓢进行运行一下。
GitHub网址:https://github.com/cxli233/ggpathway
**获得本教程代码链接:**任意赞赏即可获得
绘图
1, 导入所需R包
library(tidyverse)
library(igraph)
library(ggraph)
library(readxl)
library(viridis)
library(RColorBrewer)
#BiocManager::install("rcartocolor")
library(rcartocolor)
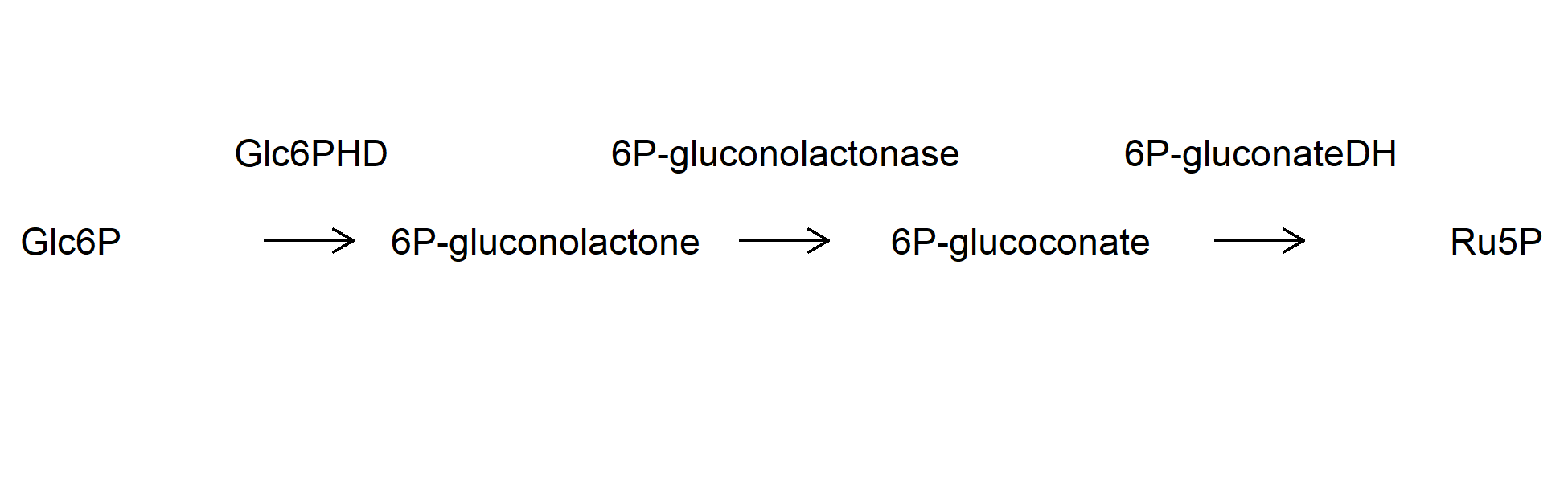
exaple one
- 创建数据
### Edge table
example1_edge_table <- tribble(
~from, ~to, ~label,
"Glc6P", "6P-gluconolactone", "Glc6PHD",
"6P-gluconolactone", "6P-glucoconate", "6P-gluconolactonase",
"6P-glucoconate", "Ru5P", "6P-gluconateDH"
)
> head(example1_edge_table)
# A tibble: 3 × 3
from to label
<chr> <chr> <chr>
1 Glc6P 6P-gluconolactone Glc6PHD
2 6P-gluconolactone 6P-glucoconate 6P-gluconolactonase
3 6P-glucoconate Ru5P 6P-gluconateDH
### Node table
example1_nodes_table <- tribble(
~name, ~x, ~y,
"Glc6P", 1, 0,
"6P-gluconolactone", 2, 0,
"6P-glucoconate", 3, 0,
"Ru5P", 4, 0
)
Make network object and graph
## Make network object and graph
example1_network <- graph_from_data_frame(
d = example1_edge_table,
vertices = example1_nodes_table,
directed = T
)
head(example1_nodes_table)
> head(example1_nodes_table)
# A tibble: 4 × 3
name x y
<chr> <dbl> <dbl>
1 Glc6P 1 0
2 6P-gluconolactone 2 0
3 6P-glucoconate 3 0
4 Ru5P 4 0
绘图
ggraph(example1_network, layout = "manual",
x = x, y = y) +
geom_node_text(aes(label = name), hjust = 0.5) +
geom_edge_link(aes(label = example1_edge_table$label),
angle_calc = 'along',
label_dodge = unit(2, 'lines'),
arrow = arrow(length = unit(0.5, 'lines')),
start_cap = circle(4, 'lines'),
end_cap = circle(4, 'lines')) +
theme_void()
ggsave("../Results/Pentose_1.svg", height = 2, width = 6.5, bg = "white")
ggsave("../Results/Pentose_1.png", height = 2, width = 6.5, bg = "white")
Example 2: more complex pathway
- 导入所需数据
setwd("E:\\小杜的生信筆記\\2023\\20231022_ggpathway")
##'@input data
example2_edges <- read_excel("Data/OPPP_edges.xlsx")
example2_nodes <- read_excel("Data/OPPP_nodes.xlsx")
head(example2_edges)
# A tibble: 6 × 3
from to label
<chr> <chr> <chr>
1 Glc6P 6P-gluconolactone Glc6PHD
2 6P-gluconolactone 6P-glucoconate 6P-gluconolactonase
3 6P-glucoconate Ru5P 6P-gluconateDH
4 Ru5P R5P R5P isomerase
5 Ru5P Xu5P_1 R5P epimerase
6 Xu5P_1 R5P T
head(example2_nodes)
# A tibble: 6 × 5
name x y carbons label
<chr> <dbl> <dbl> <dbl> <chr>
1 Glc6P 0 0 6 Glc6P
2 6P-gluconolactone 0 -1 6 6P-gluconolactone
3 6P-glucoconate 0 -2 6 6P-glucoconate
4 Ru5P 0 -3 5 Ru5P
5 R5P -0.5 -4 5 R5P
6 Xu5P_1 0 -4 5 Xu5P
- 计算 nodes
example2_nodes <- example2_nodes %>%
mutate(label = str_remove(name, "_\\d"))
head(example2_nodes)
# A tibble: 6 × 5
name x y carbons label
<chr> <dbl> <dbl> <dbl> <chr>
1 Glc6P 0 0 6 Glc6P
2 6P-gluconolactone 0 -1 6 6P-gluconolactone
3 6P-glucoconate 0 -2 6 6P-glucoconate
4 Ru5P 0 -3 5 Ru5P
5 R5P -0.5 -4 5 R5P
6 Xu5P_1 0 -4 5 Xu5P
example2_network <- graph_from_data_frame(
d = example2_edges,
vertices = example2_nodes,
directed = T
)
- 绘图
ggraph(example2_network, layout = "kk") +
geom_node_point(size = 3, aes(fill = as.factor(carbons)),
alpha = 0.8, shape = 21, color = "grey20") +
geom_node_text(aes(label = label), hjust = 0.5, repel = T) +
geom_edge_link(#aes(label = example2_edges$label),
#angle_calc = 'along',
label_dodge = unit(2, 'lines'),
arrow = arrow(length = unit(0.4, 'lines')),
start_cap = circle(1, 'lines'),
end_cap = circle(2, 'lines')) +
scale_fill_manual(values = carto_pal(7, "Vivid")) +
labs(fill = "Carbons") +
theme_void()
ggsave("Results/Pentose_2.svg", height = 5, width = 4, bg = "white")
ggsave("Results/Pentose_2.png", height = 5, width = 4, bg = "white")

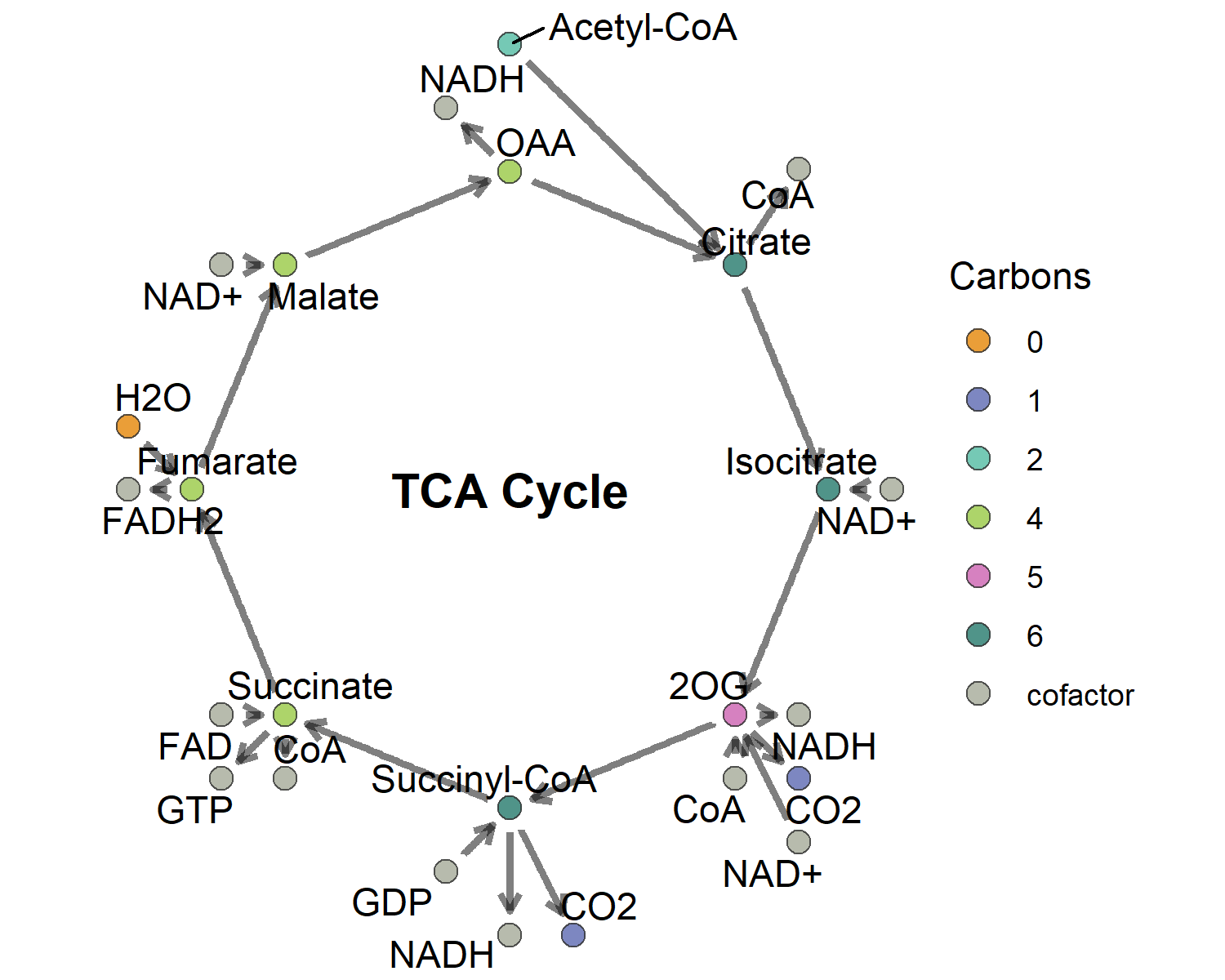
Example 3: circular pathway
- 导入数据和数据转换
example3_edges <- read_excel("Data/TCA_cycle_edges.xlsx")
example3_nodes <- read_excel("Data/TCA_cycle_nodes.xlsx")
head(example3_edges)
head(example3_nodes)
example3_nodes <- example3_nodes %>%
mutate(label = str_remove(name, "_\\d"))
head(example3_nodes)
example3_network <- graph_from_data_frame(
d = example3_edges,
vertices = example3_nodes,
directed = T
)
- 绘图
ggraph(example3_network, layout = "manual",
x = x, y = y) +
geom_node_point(size = 3, aes(fill = as.factor(carbons)),
alpha = 0.8, shape = 21, color = "grey20") +
geom_edge_link(arrow = arrow(length = unit(0.4, 'lines')),
start_cap = circle(0.5, 'lines'),
end_cap = circle(0.5, 'lines'),
width = 1.1, alpha = 0.5) +
geom_node_text(aes(label = label), hjust = 0.5, repel = T) +
annotate(geom = "text", label = "TCA Cycle",
x = 0, y = 0, size = 5, fontface = "bold") +
scale_fill_manual(values = carto_pal(7, "Vivid")) +
labs(fill = "Carbons") +
theme_void() +
coord_fixed()
Subsetting pathway
- 数据和计算转换
example3_nodes_trim <- example3_nodes %>%
filter(carbons != "cofactor")
example3_edges_trim <- example3_edges %>%
filter(from %in% example3_nodes_trim$name &
to %in% example3_nodes_trim$name)
example3_network_trim <- graph_from_data_frame(
d = example3_edges_trim,
vertices = example3_nodes_trim,
directed = T
)
绘图
ggraph(example3_network_trim, layout = "manual",
x = x, y = y) +
geom_node_point(size = 3, aes(fill = as.factor(carbons)),
alpha = 0.8, shape = 21, color = "grey20") +
geom_edge_link(arrow = arrow(length = unit(0.4, 'lines')),
start_cap = circle(0.5, 'lines'),
end_cap = circle(1, 'lines'),
width = 1.1, alpha = 0.5) +
geom_node_text(aes(label = label), hjust = 0.5, repel = T) +
annotate(geom = "text", label = "TCA Cycle",
x = 0, y = 0, size = 5, fontface = "bold") +
scale_fill_manual(values = carto_pal(7, "Vivid")) +
labs(fill = "Carbons") +
theme_void() +
coord_fixed()
ggsave("Results/TCA_2.svg", height = 4, width = 5, bg = "white")
ggsave("Results/TCA_2.png", height = 4, width = 5, bg = "white")

往期文章:
1. 复现SCI文章系列专栏
2. 《生信知识库订阅须知》,同步更新,易于搜索与管理。

3. 最全WGCNA教程(替换数据即可出全部结果与图形)
4. 精美图形绘制教程
5. 转录组分析教程
小杜的生信筆記 ,主要发表或收录生物信息学的教程,以及基于R的分析和可视化(包括数据分析,图形绘制等);分享感兴趣的文献和学习资料!!























 998
998











 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










