基因调控网络,当前研究如火如荼,为疾病的诊断提供靶向参考意义。深度学习已经超越传统方法,像经典的DeepSEM,来个开端。MetaSEM不行,还有CNN,Transformer。这些还缺乏一个下游。当然SCENIC是较好的下游工具。还是没像SCENIC+考虑的多组学那么有用。scRNA-seq勉强够水文章吧。那今天就来用一下SCENIC.
SCENIC调控推断安装过程。
安装R环境,我的是新安装,之前安装的arrow包安装不上。换一个环境Windows看看,不至于吊死在一个地方吧。window就可以安装成功:
这是我的R最新版本:
Download R-4.3.3 for Windows;
接下来直接运行bin下的R.exe安装。
参考官网如下:
http://htmlpreview.github.io/?https://github.com/aertslab/SCENIC/blob/master/inst/doc/SCENIC_Setup.html
(1)安装AUCell,RcisTarget
安装:AUCell,RcisTarget;如果安装不成功,直接下载二进制zip(当然按照他的RcisTarget安装不成功,需要手动下载二进制安装,会提升安装依赖包arrow,使用BiocManager安装即可),在用library()导入当前包,查看需要安装哪些依赖包,逐一安装即可。
(2)安装:SCENIC
devtools::install_github(“aertslab/SCENIC”)
但一般安装不成功,那就手动,直接从github下载SCENIC-master.zip,解压SCENIC-master。
使用如下安装:
devtools::install("SCENIC-master")
#当然会出错,缺失依赖包,安装即可,
如我的缺失:BiocManager::install('ggrepel')
如直接安装出错:
devtools::install_github("aertslab/SCopeLoomR", build_vignettes = TRUE)
Error: Failed to install 'unknown package' from GitHub:
Timeout was reached: [api.github.com] Failed to connect to api.github.com port 443 after 10005 ms: Timeout was reached
In addition: Warning messages:
1: In readChar(path, nchars = file.info(path)$size, ...) :
can only read in bytes in a non-UTF-8 MBCS locale
2: In readChar(path, nchars = file.info(path)$size, ...) :
can only read in bytes in a non-UTF-8 MBCS locale
3: In readChar(path, nchars = file.info(path)$size, ...) :
can only read in bytes in a non-UTF-8 MBCS locale
手动到github下载SCopeLoomR-master.zip,解压。安装
devtools::install("D:/soft_install/R-4.3.3/lry_temp/SCopeLoomR-master")
这样就安装成功了:* DONE (SCopeLoomR)
安装成功看版本如下:说明安装成功。
> packageDescription("AUCell")
Package: AUCell
Type: Package
Title: AUCell: Analysis of 'gene set' activity in single-cell RNA-seq
data (e.g. identify cells with specific gene signatures)
Version: 1.24.0
Date: 2022-12-01
> packageDescription("RcisTarget")
Package: RcisTarget
Type: Package
Title: RcisTarget Identify transcription factor binding motifs enriched
on a list of genes or genomic regions
Version: 1.22.0
Date: 2022-12-01
> packageDescription("SCENIC")
Package: SCENIC
Type: Package
Title: SCENIC (Single Cell rEgulatory Network Inference and Clustering)
Version: 1.3.1
Date: 2022-05-02
下面是之前linux安装不成功的记录,不用看,死活安装不成功arrow,那就换window吧。跳过。
安装前,需要启动我的R环境,我之前安装服务器启动如下,然后输入R,就可以运行R了。
conda activate lry_R4.2.2
参考官网连接:
http://htmlpreview.github.io/?https://github.com/aertslab/SCENIC/blob/master/inst/doc/SCENIC_Setup.html
(1)安装必要的包。
BiocManager::install(c("AUCell", "RcisTarget"))
安装RcisTarget不成功,报错arrow包安装出错,如下。
Error: package or namespace load failed for ‘arrow’ in dyn.load(file, DLLpath = DLLpath, ...)
或者什么状态0,各种安装失败
解决方案:手动下载x86_64版本,二进制版本,自行安装。
下载地址:
https://mirrors.sjtug.sjtu.edu.cn/cran/web/packages/arrow/index.html
package_path1 <-"/home/jx-vmlab/下载/arrow_15.0.1.tgz"
install.packages(package_path1,repos=NULL)
直到这里
我这里没有安装他的共表达推断网络(不过他给我自动安装了。不用就行):如,GENIE3,因为他的共表达网络效果不好,我要替换我自己的共表达网络,只是用他后面的来进一步完成我的可视化实验。
(3)物种特异性数据库,需要安装,老鼠,人。
去到官网下载:scenic给的地址是旧的,不对应motifs的更新,不能用,要用只能下载老的old
如新的,是这样的,命名。暂不用
https://resources.aertslab.org/cistarget/databases/mus_musculus/mm9/refseq_r45/mc9nr/gene_based/mm9-500bp-upstream-7species.mc9nr.genes_vs_motifs.rankings.feather
https://resources.aertslab.org/cistarget/databases/homo_sapiens/hg19/refseq_r45/mc9nr/gene_based/hg19-500bp-upstream-7species.mc9nr.genes_vs_motifs.rankings.feather
注意下载这个旧的后面才能用,要不然报错
Error in dplyr::pull():
! Can’t extract columns that don’t exist.
x Column features doesn’t exist.
这个是旧的下载地址。
老鼠
https://resources.aertslab.org/cistarget/databases/old/mus_musculus/mm9/refseq_r45/mc9nr/gene_based/mm9-500bp-upstream-7species.mc9nr.feather
https://resources.aertslab.org/cistarget/databases/old/mus_musculus/mm9/refseq_r45/mc9nr/gene_based/mm9-tss-centered-10kb-7species.mc9nr.feather
人
https://resources.aertslab.org/cistarget/databases/old/homo_sapiens/hg19/refseq_r45/mc9nr/gene_based/hg19-500bp-upstream-7species.mc9nr.feather
https://resources.aertslab.org/cistarget/databases/old/homo_sapiens/hg19/refseq_r45/mc9nr/gene_based/hg19-tss-centered-10kb-7species.mc9nr.feather
将下载的文件放到对应位置,即可。如我的都放这:lry_temp
cisTarget_databases<-c('D:/soft_install/R-4.3.3/lry_temp/')
library(RcisTarget)
data(list="motifAnnotations_mgi_v9", package="RcisTarget")
motifAnnotations_mgi <- motifAnnotations_mgi_v9
scenicOptions <- initializeScenic(org="mgi", dbDir=cisTarget_databases, nCores=10)
初始成功,提示如下:
Motif databases selected:
mm9-500bp-upstream-7species.mc9nr.feather
mm9-tss-centered-10kb-7species.mc9nr.feather
Using the column 'features' as feature index for the ranking database.
Using the column 'features' as feature index for the ranking database.
(4)接下来用的共表达网络来进行下游分析。
先用他的来跑跑效果:参考官网:
http://htmlpreview.github.io/?https://github.com/aertslab/SCENIC/blob/master/inst/doc/SCENIC_Running.html
生成一系列对应文件,在int文件夹下。最终热图不错。
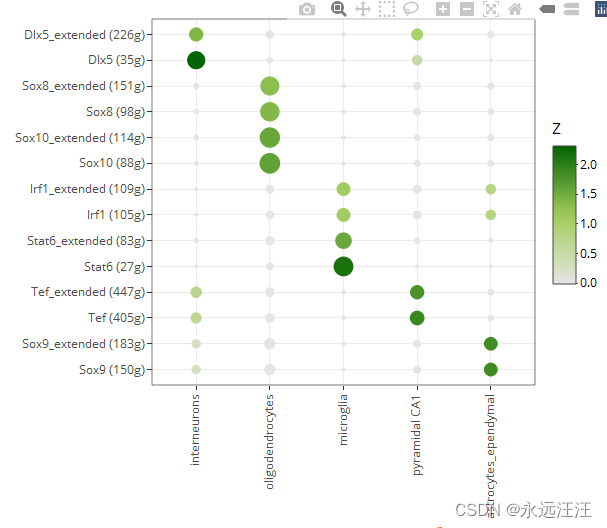
上面运行得到最终热图所有代码如下
### Load data
rm(list = ls())
library('SCENIC')
loomPath <- system.file(package="SCENIC", "examples/mouseBrain_toy.loom")
library(SCopeLoomR)
loom <- open_loom(loomPath)
exprMat <- get_dgem(loom)
cellInfo <- get_cell_annotation(loom)
close_loom(loom)
# dbFiles <- c("https://resources.aertslab.org/cistarget/databases/mus_musculus/mm9/refseq_r45/mc9nr/gene_based/mm9-500bp-upstream-7species.mc9nr.feather",
# "https://resources.aertslab.org/cistarget/databases/mus_musculus/mm9/refseq_r45/mc9nr/gene_based/mm9-tss-centered-10kb-7species.mc9nr.feather")
# cisTarget_databases<-c('D:/soft_install/R-4.3.3/lry_temp/mm9-500bp-upstream-7species.mc9nr.genes_vs_motifs.rankings.feather',
# 'D:/soft_install/R-4.3.3/lry_temp/mm9-tss-centered-10kb-7species.mc9nr.genes_vs_motifs.rankings.feather')
cisTarget_databases<-c('D:/soft_install/R-4.3.3/lry_temp/')
library(RcisTarget)
data(list="motifAnnotations_mgi_v9", package="RcisTarget")
motifAnnotations_mgi <- motifAnnotations_mgi_v9
scenicOptions <- initializeScenic(org="mgi", dbDir=cisTarget_databases, nCores=10)
# scenicOptions@inputDatasetInfo$cellInfo <- "int/cellInfo.Rds"
saveRDS(scenicOptions, file="int/scenicOptions.Rds")
#862个基因,过滤剩余770
### Co-expression network
genesKept <- geneFiltering(exprMat, scenicOptions)
exprMat_filtered <- exprMat[genesKept, ]
#技术spearman,#值对称的,对角为1;产生文件1.2
runCorrelation(exprMat_filtered, scenicOptions)
exprMat_filtered_log <- log2(exprMat_filtered+1)
#运行这个产生文件1.3开头的10个文件,1.4一个文件,就是我说的TF,Target,weight
runGenie3(exprMat_filtered_log, scenicOptions)
# > head(`1.4_GENIE3_linkList`)
# TF Target weight
# 1 Sox8 Pdk4 0.8743810
# 2 Sox9 Fgfr3 0.8307635
# 3 Dlx1 Dlx5 0.8064137
# 4 Dlx5 Dlx1 0.7985320
# 5 Sox9 Slc9a3r1 0.7944334
# 6 Sox9 Gja1 0.7842589
### Build and score the GRN
exprMat_log <- log2(exprMat+1)
scenicOptions@settings$dbs <- scenicOptions@settings$dbs["10kb"] # Toy run settings
scenicOptions <- runSCENIC_1_coexNetwork2modules(scenicOptions)#产生1.5,1.6
#结果如下
# 16:08 Creating TF modules
# Number of links between TFs and targets (weight>=0.001): 6139
# [,1]
# nTFs 8
# nTargets 770
# nGeneSets 47
# nLinks 17308
#产生2.1,2.2,2.3,会出错,重启R解决;产生2.5,2.6
scenicOptions <- runSCENIC_2_createRegulons(scenicOptions, coexMethod=c("top5perTarget")) # Toy run settings
#报错下面,重启Rstudio就解决
# Error in socketAccept(socket = socket, blocking = TRUE, open = "a+b", :
# all connections are in use
#运行下面需要安装这个包
#install.packages("doMC", repos="http://R-Forge.R-project.org")
scenicOptions <- runSCENIC_3_scoreCells(scenicOptions, exprMat_log)
# Optional: Binarize activity
# aucellApp <- plotTsne_AUCellApp(scenicOptions, exprMat_log)
# savedSelections <- shiny::runApp(aucellApp)
# newThresholds <- savedSelections$thresholds
# scenicOptions@fileNames$int["aucell_thresholds",1] <- "int/newThresholds.Rds"
# saveRDS(newThresholds, file=getIntName(scenicOptions, "aucell_thresholds"))
scenicOptions <- runSCENIC_4_aucell_binarize(scenicOptions)
tsneAUC(scenicOptions, aucType="AUC") # choose settings
# Export:
# saveRDS(cellInfo, file=getDatasetInfo(scenicOptions, "cellInfo")) # Temporary, to add to loom
export2loom(scenicOptions, exprMat)
# To save the current status, or any changes in settings, save the object again:
saveRDS(scenicOptions, file="int/scenicOptions.Rds")
### Exploring output
# Check files in folder 'output'
# Browse the output .loom file @ http://scope.aertslab.org
# output/Step2_MotifEnrichment_preview.html in detail/subset:
motifEnrichment_selfMotifs_wGenes <- loadInt(scenicOptions, "motifEnrichment_selfMotifs_wGenes")
tableSubset <- motifEnrichment_selfMotifs_wGenes[highlightedTFs=="Sox8"]
viewMotifs(tableSubset)
# output/Step2_regulonTargetsInfo.tsv in detail:
regulonTargetsInfo <- loadInt(scenicOptions, "regulonTargetsInfo")
tableSubset <- regulonTargetsInfo[TF=="Stat6" & highConfAnnot==TRUE]
viewMotifs(tableSubset) #但这个看不到motiff图,问号
# Cell-type specific regulators (RSS):
regulonAUC <- loadInt(scenicOptions, "aucell_regulonAUC")
library('BiocParallel')
rss <- calcRSS(AUC=getAUC(regulonAUC), cellAnnotation=cellInfo[colnames(regulonAUC), "CellType"], )
rssPlot <- plotRSS(rss)
plotly::ggplotly(rssPlot$plot)#这个热图就好看多了,是我要的





















 6859
6859

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








