library(ggplot2)
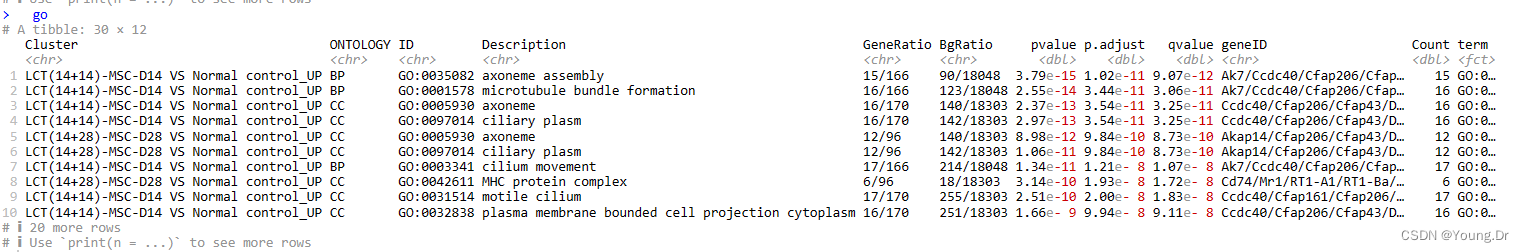
go <- xx@compareClusterResult
head(go)
go$term <- paste(go$ID, go$Description, sep = ': ')
# go <- go[order(go$ONTOLOGY, go$p.adjust, decreasing = c(TRUE, TRUE)), ]
go <- go[order( go$p.adjust, decreasing = c( FALSE) ), ]
head(go)
go$term <- factor(go$term, levels = unique(go$term))
print(dim(go))
#library(tidyr)
# install.packages("BiocManager")
# library(BiocManager)
# BiocManager::install("dplyr")
head(go)
go <-go %>%
group_by(ONTOLOGY) %>%
mutate(group_size = n()) %>%
filter(row_number() <= ifelse(group_size > 10, 10, group_size)) %>%
ungroup() %>%
select(-group_size)
go
p3=ggplot(go, aes(term, -log10(p.adjust))) +
geom_col(aes(fill = ONTOLOGY), width = 0.5, show.legend = FALSE) +
scale_fill_manual(values = c('#D06660', '#5AAD36', '#6C85F5')) +
facet_grid(ONTOLOGY~., scale = 'free_y', space = 'free_y') +
theme(panel.grid = element_blank(), panel.background = element_rect(color = 'black', fill = 'transparent')) +
scale_y_continuous(expand = expansion(mult = c(0, 0.1))) +
coord_flip() +
labs(x = '', y = '-Log10 P-Value\n') 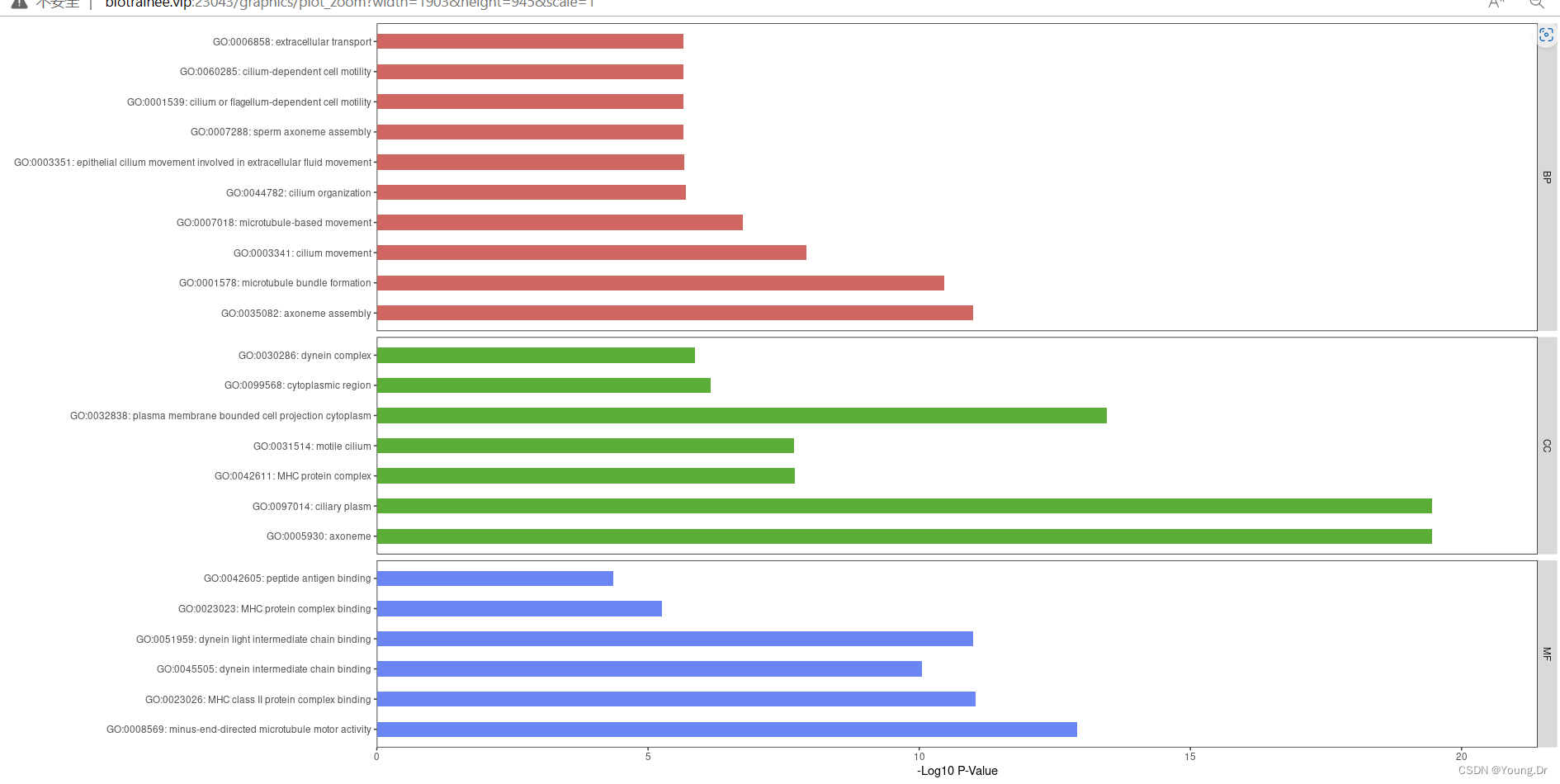
#https://mp.weixin.qq.com/s/WyT-7yKB9YKkZjjyraZdPg { library(clusterProfiler) # library(org.Hs.eg.db) # BiocManager::install("org.Rn.eg.db") # install.packages("org.Rn.eg.db") library(org.Rn.eg.db) library(ggplot2) # degs_for_nlung_vs_tlung$gene=rownames(degs_for_nlung_vs_tlung) sce.markers=df head(sce.markers) ids=bitr(sce.markers$gene,'SYMBOL','ENTREZID','org.Rn.eg.db') head(ids) sce.markers=merge(sce.markers,ids,by.x='gene',by.y='SYMBOL') head(sce.markers) sce.markers=sce.markers[sce.markers$change!="NOT",] dim(sce.markers) head(sce.markers) sce.markers$cluster=sce.markers$mygroup gcSample=split(sce.markers$ENTREZID, sce.markers$cluster) gcSample # entrez id , compareCluster #https://zhuanlan.zhihu.com/p/561522453 xx <- compareCluster(gcSample, fun="enrichGO",OrgDb="org.Rn.eg.db", readable=TRUE, ont = 'ALL', #GO Ontology,可选 BP、MF、CC,也可以指定 ALL 同时计算 3 者 pvalueCutoff=0.05) #organism="hsa", #'org.Hs.eg.db', p=dotplot(xx) p2=p+ theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=0.5)) p2 ggsave('degs_compareCluster-GO_enrichment-2.pdf',plot = p2,width = 6,height = 12,limitsize = F) xx openxlsx::write.xlsx(xx,file = "compareCluster-GO_enrichment.xlsx") } getwd() setwd("/home/data/t040413/wpx/wpx_transcriptomics_proteinomics_Metabolomics/transcriptomics/4_d28_vs_d14_treatment_effective/")

























 1408
1408

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?










