简 介
scRNA-seq数据解释被称为双链的技术产物所混淆,即代表多个细胞的单细胞转录组数据。此外,scRNA-seq细胞通量被有目的地限制,以最小化双偶体形成率。通过识别与模拟双链具有相同表达特征的细胞,DoubletFinder可以检测到许多真正的双链,并减轻了这两个限制。

原 理
DoubletFinder的原理是从现有的矩阵的细胞中根据我们预先定义好的细胞类型模拟一些双细胞出来(比如单核和T细胞的双细胞、B细胞和中性粒细胞的双细胞等等),将模拟出的双细胞和原有矩阵的细胞混合在一起,进行降维聚类,原则上人工模拟的doublets会与真实的doublets距离较近,因此计算每个细胞K最近邻细胞中人工模拟doublets的比例( pAN ),就可以根据pANN值对每个barcode的doublets概率进行排序。另外依据泊松分布的统计原理可以计算每个样本中doublets的数量,结合之前的细胞pANN值排序,就可以过滤doublets了。

步骤
DoubletFinder可分为4个步骤:
(1)从已有的scRNA-seq数据中生成人工双元;
(2)对合并的真实人工数据进行预处理;
(3)进行主成分分析,利用PC距离矩阵求出每个单元的人工k个最近邻(pANN)的比例;
(4)根据期望的双元数量排序和阈值pANN值。

软件包安装
软件包安装很简单,github模式安装即可:
## 载入需要的程辑包:DoubletFinder
if(!require(DoubletFinder))
remotes::install_github('chris-mcginnis-ucsf/DoubletFinder')数据读取
这里我们使用 pbmc3k 的数据集来做例子,完成整个分析过程,这里我们需要调用 Seurat 软件包来完成数据的处理工作。数据读入如下:
library(DoubletFinder)
library(tidyverse)
library(Seurat)
library(patchwork)
## Pre-process Seurat object (standard)
## --------------------------------------------------------------------------------------
pbmc.data <- Read10X(data.dir = "./filtered_gene_bc_matrices/hg19")
pbmc <- CreateSeuratObject(pbmc.data, project = "pbmc3k")实例操作
Step1: 数据预处理:单细胞数据质控、标准化、降维聚类等
首先这个R包的输入是经过预处理(包括归一化、降维,但不一定要聚类)的 Seurat 对象。这里pbmc是我的Seurat对象名称你可以换成你的样本对象名称。
pbmc <- NormalizeData(pbmc)
pbmc <- FindVariableFeatures(pbmc, selection.method = "vst", nfeatures = 2000)
pbmc <- ScaleData(pbmc)
pbmc <- RunPCA(pbmc)
pbmc <- RunUMAP(pbmc, dims = 1:10)
pbmc <- FindNeighbors(pbmc, dims = 1:10)
pbmc <- FindClusters(pbmc, resolution = 0.5)
## Modularity Optimizer version 1.3.0 by Ludo Waltman and Nees Jan van Eck
##
## Number of nodes: 2700
## Number of edges: 98199
##
## Running Louvain algorithm...
## Maximum modularity in 10 random starts: 0.8697
## Number of communities: 10
## Elapsed time: 0 seconds
print(pbmc[["pca"]], dims = 1:10)
## PC_ 1
## Positive: CST3, TYROBP, LST1, AIF1, FTL, FCN1, LYZ, FTH1, S100A9, FCER1G
## TYMP, CFD, LGALS1, S100A8, CTSS, LGALS2, SERPINA1, SPI1, IFITM3, PSAP
## Negative: MALAT1, LTB, IL32, CD2, ACAP1, STK17A, CTSW, CD247, CCL5, GIMAP5
## AQP3, GZMA, CST7, TRAF3IP3, MAL, HOPX, ITM2A, GZMK, MYC, GIMAP7
## PC_ 2
## Positive: NKG7, PRF1, CST7, GZMA, GZMB, FGFBP2, CTSW, GNLY, GZMH, SPON2
## CCL4, FCGR3A, CCL5, CD247, XCL2, CLIC3, AKR1C3, SRGN, HOPX, CTSC
## Negative: CD79A, MS4A1, TCL1A, HLA-DQA1, HLA-DRA, HLA-DQB1, LINC00926, CD79B, HLA-DRB1, CD74
## HLA-DPB1, HLA-DMA, HLA-DQA2, HLA-DRB5, HLA-DPA1, HLA-DMB, FCRLA, HVCN1, LTB, BLNK
## PC_ 3
## Positive: HLA-DQA1, CD79A, CD79B, HLA-DQB1, HLA-DPB1, CD74, HLA-DPA1, MS4A1, HLA-DRB1, HLA-DRB5
## HLA-DRA, HLA-DQA2, TCL1A, LINC00926, HLA-DMB, HLA-DMA, HVCN1, FCRLA, IRF8, BLNK
## Negative: PPBP, PF4, SDPR, SPARC, GNG11, NRGN, GP9, RGS18, TUBB1, CLU
## HIST1H2AC, AP001189.4, ITGA2B, CD9, TMEM40, CA2, PTCRA, ACRBP, MMD, NGFRAP1
## PC_ 4
## Positive: HLA-DQA1, HIST1H2AC, PF4, CD79A, SDPR, CD79B, PPBP, GNG11, HLA-DQB1, SPARC
## MS4A1, CD74, GP9, HLA-DPB1, RGS18, NRGN, PTCRA, CD9, HLA-DQA2, AP001189.4
## Negative: VIM, S100A8, S100A6, S100A4, TMSB10, S100A9, IL32, GIMAP7, S100A10, LGALS2
## RBP7, MAL, FCN1, LYZ, CD2, S100A12, MS4A6A, FYB, S100A11, AQP3
## PC_ 5
## Positive: LTB, VIM, AQP3, PPA1, MAL, KIAA0101, CD2, CORO1B, CYTIP, FYB
## IL32, TRADD, ANXA5, TUBA1B, HN1, PTGES3, TYMS, ITM2A, COTL1, GPR183
## Negative: GZMB, FGFBP2, NKG7, GNLY, PRF1, CCL4, CST7, SPON2, GZMA, GZMH
## CLIC3, XCL2, CTSW, TTC38, AKR1C3, CCL5, IGFBP7, XCL1, S100A8, CCL3
## PC_ 6
## Positive: TYMS, KIAA0101, TK1, RRM2, S100A8, MKI67, MYBL2, ZWINT, BIRC5, GINS2
## LGALS2, MS4A6A, GSTP1, GPX1, KIFC1, FOLR3, STMN1, GAPDH, ASF1B, S100A12
## Negative: FCGR3A, CKB, MS4A7, HMOX1, IFITM2, MALAT1, VMO1, CTD-2006K23.1, ABI3, PPM1N
## IFITM3, ADA, LYN, LILRB2, WARS, C1QA, TIMP1, LYPD2, CEBPB, NAAA
## PC_ 7
## Positive: TYMS, KIAA0101, ZWINT, RRM2, TK1, GINS2, BIRC5, MKI67, FCGR3A, CKB
## KIFC1, AURKB, CLSPN, MYBL2, MS4A7, NUSAP1, RAD51, ASF1B, CCNA2, TOP2A
## Negative: FCER1A, CLEC10A, SERPINF1, CD1C, CACNA2D3, ID1, ENHO, ALDH2, VIM, GPX1
## HLA-DQA2, ANXA1, LGALS2, VAMP8, HLA-DQB1, HLA-DMB, HLA-DQA1, RGS1, ETHE1, HLA-DMA
## PC_ 8
## Positive: AKR1C3, IGFBP7, SPON2, GZMB, TMSB10, CLIC3, FGFBP2, HAVCR2, GNLY, S100B
## MAL, TRABD2A, TTC38, NGFRAP1, KIR2DL3, C1orf162, UBE2F, DAB2, CD247, FCER1G
## Negative: GZMK, CCL5, KLRG1, LYAR, IL32, GZMA, S100A4, NKG7, CST7, JAKMIP1
## TIGIT, NCR3, GYG1, APOBEC3G, PRR5, S100A10, S100A6, SYNE2, GPR171, HLA-DPB1
## PC_ 9
## Positive: RBP7, S100A12, S100A8, FOLR3, LINC00926, CD79A, S100A9, LTB, IFI35, AIM2
## GUCD1, CD79B, MS4A1, TREX1, CDA, CD82, NFE2, NFATC1, KIAA0125, RETN
## Negative: FCER1A, CLEC10A, ENHO, CD1C, SERPINF1, RP6-91H8.3, CACNA2D3, PPP2R3C, FABP5, HLA-DQA2
## TUBA1B, ABI3, PLD4, IL1B, GAS6, HLA-DQA1, HLA-DPA1, PHACTR1, HLA-DPB1, LILRB2
## PC_ 10
## Positive: IGJ, MZB1, AQP3, GAPDH, AL928768.3, S100A10, ADA, CWC27, NOP58, ECHDC1
## SMPDL3A, TNFRSF13B, VIM, CRIP2, CKB, PPIB, ARL2, ACTB, FLNA, PSMC4
## Negative: IFIT1, GBP1, ISG15, MX1, IFI6, UBE2L6, TNFSF10, IFIT2, GIMAP4, MX2
## TREX1, APOBEC3B, GMPR, APOBEC3A, IFI35, ZBP1, IFI44, IRF7, LYAR, GIMAP7
head(pbmc$seurat_clusters, 5)
## AAACATACAACCAC-1 AAACATTGAGCTAC-1 AAACATTGATCAGC-1 AAACCGTGCTTCCG-1
## 2 3 2 5
## AAACCGTGTATGCG-1
## 6
## Levels: 0 1 2 3 4 5 6 7 8 9
g1 <- DimPlot(pbmc, reduction = "umap")
g1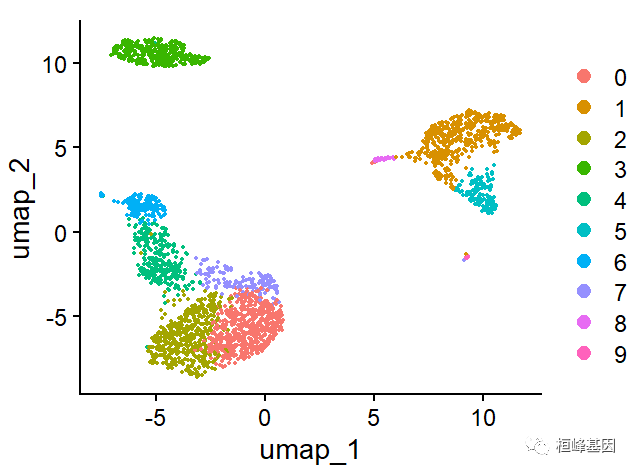
参数说明:
DoubletFinder takes the following arguments:
seu ~ This is a fully-processed Seurat object (i.e., after NormalizeData, FindVariableGenes, ScaleData, RunPCA, and RunTSNE have all been run).
PCs ~ The number of statistically-significant principal components, specified as a range (e.g., PCs = 1:10)
pN ~ This defines the number of generated artificial doublets, expressed as a proportion of the merged real-artificial data. Default is set to 25%, based on observation that DoubletFinder performance is largely pN-invariant (see McGinnis, Murrow and Gartner 2019, Cell Systems).
pK ~ This defines the PC neighborhood size used to compute pANN, expressed as a proportion of the merged real-artificial data. No default is set, as pK should be adjusted for each scRNA-seq dataset. Optimal pK values should be estimated using the strategy described below.
nExp ~ This defines the pANN threshold used to make final doublet/singlet predictions. This value can best be estimated from cell loading densities into the 10X/Drop-Seq device, and adjusted according to the estimated proportion of homotypic doublets.Step2: 确定参数,寻找最优pK值
对Cell哈希和Demuxlet数据集进行pN- pk参数扫描的ROC分析表明,DoubletFinder的性能在很大程度上不受pN值选择的影响:
## pK Identification (no ground-truth)
## ---------------------------------------------------------------------------------------
sweep.res.list_pbmc <- paramSweep_v3(pbmc, PCs = 1:10, sct = FALSE)
sweep.stats_pbmc <- summarizeSweep(sweep.res.list_pbmc, GT = FALSE)
bcmvn_pbmc <- find.pK(sweep.stats_pbmc)
## NULL
mpK <- as.numeric(as.vector(bcmvn_pbmc$pK[which.max(bcmvn_pbmc$BCmetric)]))
mpK
## [1] 0.01Step3: 双细胞比例计算
对模拟scRNA-seq数据进行pN-pK参数扫描的ROC分析:
(I)细胞状态数量不同,
(II)转录异质性程度不同,
表明(I)最佳pK值选择取决于细胞状态的总数,(II)当应用于转录同质数据时,DoubletFinder的性能会受到影响。
## Homotypic Doublet Proportion Estimate
## -------------------------------------------------------------------------------------
annotations <- pbmc$seurat_clusters
head(annotations)
## AAACATACAACCAC-1 AAACATTGAGCTAC-1 AAACATTGATCAGC-1 AAACCGTGCTTCCG-1
## 2 3 2 5
## AAACCGTGTATGCG-1 AAACGCACTGGTAC-1
## 6 0
## Levels: 0 1 2 3 4 5 6 7 8 9
homotypic.prop <- modelHomotypic(annotations)
homotypic.prop
## [1] 0.1482741
nExp_poi <- round(0.075 * nrow(pbmc@meta.data)) ## Assuming 7.5% doublet formation rate - tailor for your dataset
nExp_poi
## [1] 202
nExp_poi.adj <- round(nExp_poi * (1 - homotypic.prop))
nExp_poi.adj
## [1] 172Step5: 结果可视化
这里确定了Doublet-Low Confidience, Doublet-High Confidience,Singlet三种细胞类型。
## [1] "Creating 900 artificial doublets..."
## [1] "Creating Seurat object..."
## [1] "Normalizing Seurat object..."
## [1] "Finding variable genes..."
## [1] "Scaling data..."
## [1] "Running PCA..."
## [1] "Calculating PC distance matrix..."
## [1] "Computing pANN..."
## [1] "Classifying doublets.."
## [1] 0.05246914 0.29938272## Plot results
## ---------------------------------------------------------------------------
head(pbmc@meta.data$DF.classifications_0.25_0.09_202, 5)
## [1] "Singlet" "Singlet" "Doublet" "Singlet" "Singlet"
head(pbmc@meta.data$DF.classifications_0.25_0.09_172, 5)
## [1] "Singlet" "Singlet" "Doublet" "Singlet" "Singlet"
pbmc@meta.data[, "DF_hi.lo"] <- pbmc@meta.data$DF.classifications_0.25_0.09_202
pbmc@meta.data$DF_hi.lo[which(pbmc@meta.data$DF_hi.lo == "Doublet" & pbmc@meta.data$DF.classifications_0.25_0.09_172 ==
"Singlet")] <- "Doublet-Low Confidience"
pbmc@meta.data$DF_hi.lo[which(pbmc@meta.data$DF_hi.lo == "Doublet")] <- "Doublet-High Confidience"
table(pbmc@meta.data$DF_hi.lo)
##
## Doublet-High Confidience Doublet-Low Confidience Singlet
## 172 30 2498
## 结果展示,分类结果在pbmc@meta.data中
g2 <- DimPlot(pbmc, reduction = "umap", group.by = "DF_hi.lo", cols = c("black",
"red", "gold"))
g2
g1 | g2
Reference
McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019;8(4):329-337.e4. doi:10.1016/j.cels.2019.03.003
单细胞生信分析教程
桓峰基因公众号推出单细胞生信分析教程并配有视频在线教程,目前整理出来的相关教程目录如下:
SCS【4】单细胞转录组数据可视化分析 (Seurat 4.0)
SCS【6】单细胞转录组之细胞类型自动注释 (SingleR)
SCS【7】单细胞转录组之轨迹分析 (Monocle 3) 聚类、分类和计数细胞
SCS【8】单细胞转录组之筛选标记基因 (Monocle 3)
SCS【9】单细胞转录组之构建细胞轨迹 (Monocle 3)
SCS【10】单细胞转录组之差异表达分析 (Monocle 3)
SCS【11】单细胞ATAC-seq 可视化分析 (Cicero)
SCS【12】单细胞转录组之评估不同单细胞亚群的分化潜能 (Cytotrace)
SCS【13】单细胞转录组之识别细胞对“基因集”的响应 (AUCell)
SCS【15】细胞交互:受体-配体及其相互作用的细胞通讯数据库 (CellPhoneDB)
SCS【16】从肿瘤单细胞RNA-Seq数据中推断拷贝数变化 (inferCNV)
SCS【17】从单细胞转录组推断肿瘤的CNV和亚克隆 (copyKAT)
SCS【18】细胞交互:受体-配体及其相互作用的细胞通讯数据库 (iTALK)
SCS【21】单细胞空间转录组可视化 (Seurat V5)
SCS【22】单细胞转录组之 RNA 速度估计 (Velocyto.R)
SCS【24】单细胞数据量化代谢的计算方法 (scMetabolism)
SCS【25】单细胞细胞间通信第一部分细胞通讯可视化(CellChat)
SCS【26】单细胞细胞间通信第二部分通信网络的系统分析(CellChat)
SCS【28】单细胞转录组加权基因共表达网络分析(hdWGCNA)
SCS【29】单细胞基因富集分析 (singleseqgset)
SCS【30】单细胞空间转录组学数据库(STOmics DB)
SCS【31】减少障碍,加速单细胞研究数据库(Single Cell PORTAL)
SCS【32】基于scRNA-seq数据中推断单细胞的eQTLs (eQTLsingle)
SCS【33】单细胞转录之全自动超快速的细胞类型鉴定 (ScType)
SCS【34】单细胞/T细胞/抗体免疫库数据分析(immunarch)
利用这个软件包实现了快速去除双细胞,有需求的老师可以联系桓峰基因,关注桓峰基因公众号,轻松学生信,高效发文章!
号外号外,桓峰基因单细胞生信分析免费培训课程即将开始,快来报名吧!
桓峰基因,铸造成功的您!
未来桓峰基因公众号将不间断的推出单细胞系列生信分析教程,
敬请期待!!
桓峰基因官网正式上线,请大家多多关注,还有很多不足之处,大家多多指正!
http://www.kyohogene.com/
桓峰基因和投必得合作,文章润色优惠85折,需要文章润色的老师可以直接到网站输入领取桓峰基因专属优惠券码:KYOHOGENE,然后上传,付款时选择桓峰基因优惠券即可享受85折优惠哦!https://www.topeditsci.com/

























 303
303

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








