原始教程链接:https://github.com/iMetaScience/iMetaPlot/tree/main/221108NMDS
写在前面
非度量多维尺度分析(Non-metric multidimensional scaling, NMDS),是基于相异矩阵或距离矩阵进行排序分析的间接梯度分析方法,在微生物组研究中可以用来展示群落beta多样性。本期我们挑选2022年2月24日刊登在iMeta上的Linking soil fungi to bacterial community assembly in arid ecosystems - iMeta:西农韦革宏团队焦硕等-土壤真菌驱动细菌群落的构建,选择文章的Figure 6C进行复现,基于vegan包,讲解和探讨和NMDS分析和可视化的方法,先上原图:

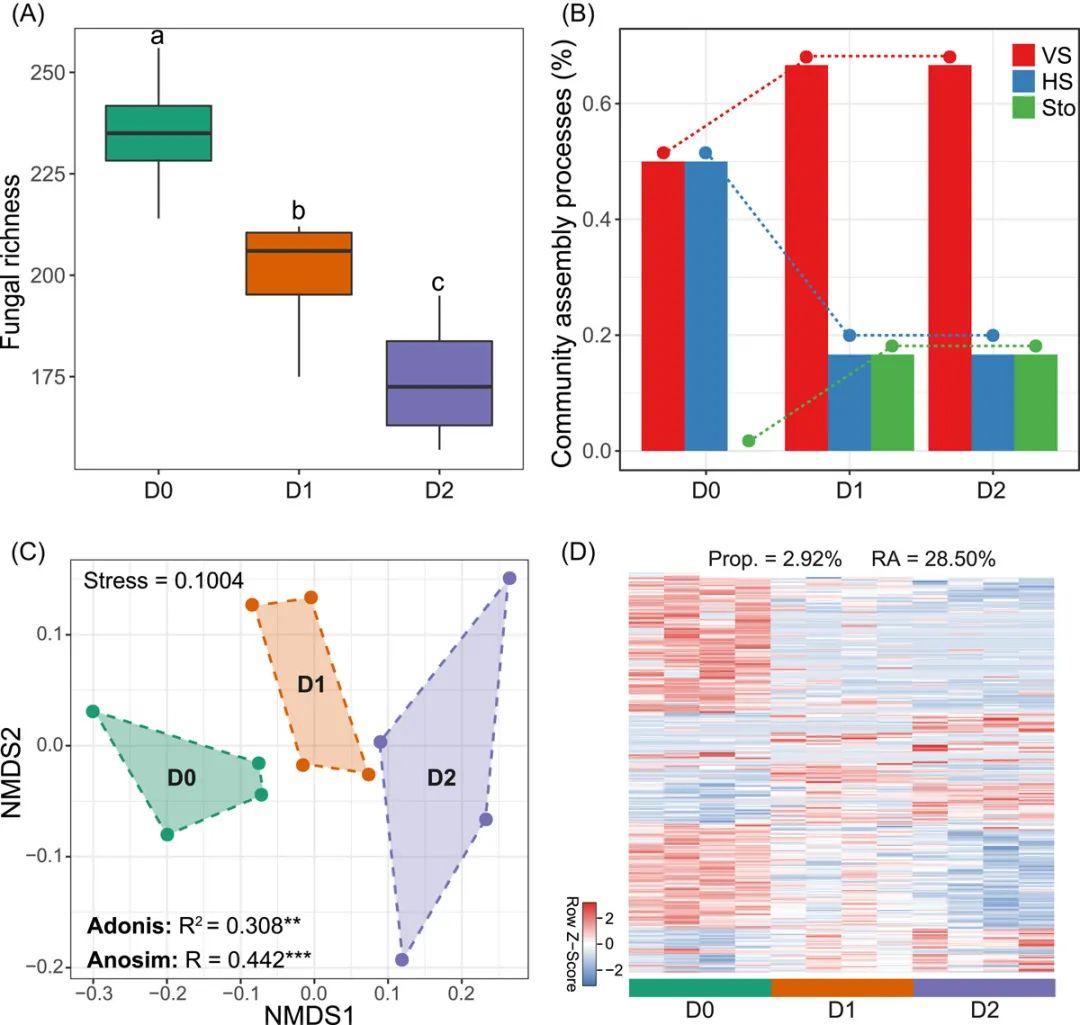
接下来,我们将通过详尽的代码逐步拆解原图,最终实现对原图的复现。代码编写及注释:农心生信工作室。
R包检测和安装
01
安装核心R包vegan以及ggplot2,并载入所有R包。
if (!require("vegan"))
install.packages('vegan')
if (!require("ggplot2"))
install.packages('ggplot2')
# 加载包
library(vegan)
library(ggplot2)生成测试数据
02
由于缺少原始数据,因此本例使用vegan包自带的dune数据集进行测试。dune包含了20个样品,每个样品有30个物种丰度,每一行是一个样品,每一列是一个物种。
# 载入dune数据集
data(dune)
#载入dune包含分组信息等的元数据(即metadata),分组信息为Management列
data(dune.env)NMDS分析
03
获取数据后,即可利用vegan包进行NMDS分析。
#计算bray_curtis距离
distance <- vegdist(dune, method = 'bray')
#NMDS排序分析,k = 2预设两个排序轴
nmds <- metaMDS(distance, k = 2)
#> Run 0 stress 0.1192678
#> Run 1 stress 0.1192678
#> ... Procrustes: rmse 4.495733e-05 max resid 0.0001375161
#> ... Similar to previous best
#> Run 2 stress 0.1183186
#> ... New best solution
#> ... Procrustes: rmse 0.02026799 max resid 0.06495211
#> Run 3 stress 0.1183186
#> ... New best solution
#> ... Procrustes: rmse 1.832694e-05 max resid 5.57604e-05
#> ... Similar to previous best
#> Run 4 stress 0.1809577
#> Run 5 stress 0.1192678
#> Run 6 stress 0.1183186
#> ... New best solution
#> ... Procrustes: rmse 5.582524e-06 max resid 1.803473e-05
#> ... Similar to previous best
#> Run 7 stress 0.1192678
#> Run 8 stress 0.1192678
#> Run 9 stress 0.1192678
#> Run 10 stress 0.1192678
#> Run 11 stress 0.1192679
#> Run 12 stress 0.1808911
#> Run 13 stress 0.1192678
#> Run 14 stress 0.1183186
#> ... Procrustes: rmse 5.943311e-06 max resid 1.823899e-05
#> ... Similar to previous best
#> Run 15 stress 0.1886532
#> Run 16 stress 0.1192678
#> Run 17 stress 0.1183186
#> ... Procrustes: rmse 3.001088e-06 max resid 9.607646e-06
#> ... Similar to previous best
#> Run 18 stress 0.1192679
#> Run 19 stress 0.1808911
#> Run 20 stress 0.1183186
#> ... Procrustes: rmse 2.027412e-05 max resid 6.520856e-05
#> ... Similar to previous best
#> *** Best solution repeated 4 times
#查看结果
#summary(nmds)04
获取可视化所需数据。
#获得应力值(stress)
stress <- nmds$stress
#将绘图数据转化为数据框
df <- as.data.frame(nmds$points)
#与分组数据合并
df <- cbind(df, dune.env)NMDS可视化
05
根据分组绘制一个最基础的散点图。
p <- ggplot(df, aes(MDS1, MDS2))+
geom_point(aes(color = Management), size = 5)
06
我们注意到,原图中,每个分组被连接成不规则的多边形并用不同颜色表示,我们可以通过ggplot2中geom_polygon()来绘制。geom_polygon()会按照数据中出现的顺序连接观测值,内部可填充颜色。
p <- ggplot(df, aes(MDS1, MDS2))+
geom_point(aes(color = Management), size = 5)+
geom_polygon(aes(x = MDS1, y = MDS2, fill = Management, group = Management, color = Management),
alpha = 0.3, linetype = "longdash", linewidth = 1.5) #通过按顺序连接观测值绘制多边形
07
由于geom_polygon()会按照数据中出现的顺序连接观测值,因此如果我们按照df自身顺序来绘制多边形,多边形会非常奇怪,没法代表不同分组。因此,我们需要预先处理df的顺序,按合理的顺序连接观测值。
df <- df[order(df$Management), ]#先按分组排序
df$Order <- c(2, 1, 3, 1, 2, 3, 4, 5, 3, 5, 1, 6, 2, 4, 1, 2, 6, 3, 5, 4)#添加一列Order,给每个分组内观测点的手动排序
df <- df[order(df$Management, df$Order), ]#按分组和Order排序
p <- ggplot(df, aes(MDS1, MDS2))+
geom_point(aes(color = Management), size = 5)+
geom_polygon(aes(x = MDS1, y = MDS2, fill = Management, group = Management, color = Management),
alpha = 0.3, linetype = "longdash", linewidth = 1.5)
08
分别进行Anosim分析(Analysis of similarities)和PERMANOVA(即adonis)检验分析。
#设置随机种子
set.seed(123)
#基于bray-curtis距离进行PERMANOVA分析
adonis <- adonis2(dune ~ Management, data = dune.env, permutations = 999, method = "bray")
#基于bray-curtis距离进行anosim分析
anosim = anosim(dune, dune.env$Management, permutations = 999, distance = "bray")09
美化图片,并用AI微调。
# 应力值stress,Adonis R2与显著性,Anosim R与显著性
stress_text <- paste("Stress =", round(stress, 4))
adonis_text <- paste(paste("Adonis =", round(adonis$R2, 2)), "**")[1]
anosim_text <- paste(paste("Anosim =", round(anosim$statistic, 2)), "**")
p <- ggplot(df, aes(MDS1, MDS2))+
geom_point(aes(color = Management), size = 5)+
geom_polygon(aes(x = MDS1, y = MDS2, fill = Management, group = Management, color = Management), alpha = 0.3, linetype = "longdash", linewidth = 1.5)+
theme(plot.margin = unit(rep(1, 4), 'lines'),
panel.border = element_rect(fill = NA, color = "black", size = 0.5, linetype = "solid"),
panel.grid = element_blank(),
panel.background = element_rect(fill = 'white'))+
guides(color = "none", fill = "none")+
ggtitle(paste(paste(stress_text, adonis_text), anosim_text))
完整代码
if (!require("vegan"))
install.packages('vegan')
if (!require("ggplot2"))
install.packages('ggplot2')
# 加载包
library(vegan)
library(ggplot2)
# 载入dune数据集
data(dune)
#载入dune包含分组信息等的元数据(即metadata),分组信息为Management列
data(dune.env)
#计算bray_curtis距离
distance <- vegdist(dune, method = 'bray')
#NMDS排序分析,k = 2预设两个排序轴
nmds <- metaMDS(distance, k = 2)
#> Run 0 stress 0.1192678
#> Run 1 stress 0.1192678
#> ... Procrustes: rmse 1.505128e-05 max resid 4.673581e-05
#> ... Similar to previous best
#> Run 2 stress 0.1192678
#> ... Procrustes: rmse 3.715749e-06 max resid 1.009651e-05
#> ... Similar to previous best
#> Run 3 stress 0.1889642
#> Run 4 stress 0.1192679
#> ... Procrustes: rmse 0.0001542849 max resid 0.0004702712
#> ... Similar to previous best
#> Run 5 stress 0.1886532
#> Run 6 stress 0.2341212
#> Run 7 stress 0.1192678
#> ... Procrustes: rmse 1.328909e-05 max resid 4.273575e-05
#> ... Similar to previous best
#> Run 8 stress 0.1886532
#> Run 9 stress 0.1192678
#> ... Procrustes: rmse 1.903819e-05 max resid 5.828243e-05
#> ... Similar to previous best
#> Run 10 stress 0.1192678
#> ... Procrustes: rmse 6.358457e-06 max resid 1.687026e-05
#> ... Similar to previous best
#> Run 11 stress 0.119268
#> ... Procrustes: rmse 5.501506e-05 max resid 0.0001605112
#> ... Similar to previous best
#> Run 12 stress 0.1192678
#> ... New best solution
#> ... Procrustes: rmse 5.074111e-06 max resid 1.393603e-05
#> ... Similar to previous best
#> Run 13 stress 0.1192678
#> ... Procrustes: rmse 3.160318e-05 max resid 9.85043e-05
#> ... Similar to previous best
#> Run 14 stress 0.1886532
#> Run 15 stress 0.2003486
#> Run 16 stress 0.2035424
#> Run 17 stress 0.1192678
#> ... Procrustes: rmse 2.440829e-05 max resid 7.079487e-05
#> ... Similar to previous best
#> Run 18 stress 0.1183186
#> ... New best solution
#> ... Procrustes: rmse 0.02027171 max resid 0.06497302
#> Run 19 stress 0.1183186
#> ... New best solution
#> ... Procrustes: rmse 3.78469e-06 max resid 9.699447e-06
#> ... Similar to previous best
#> Run 20 stress 0.1192678
#> *** Best solution repeated 1 times
#查看结果
#summary(nmds)
#获得应力值(stress)
stress <- nmds$stress
#将绘图数据转化为数据框
df <- as.data.frame(nmds$points)
#与分组数据合并
df <- cbind(df, dune.env)
df <- df[order(df$Management), ]#先按分组排序
df$Order <- c(2, 1, 3, 1, 2, 3, 4, 5, 3, 5, 1, 6, 2, 4, 1, 2, 6, 3, 5, 4)#添加一列Order,给每个分组内观测点的手动排序
df <- df[order(df$Management, df$Order), ]#按分组和Order排序
#设置随机种子
set.seed(123)
#基于bray-curtis距离进行PERMANOVA分析
adonis <- adonis2(dune ~ Management, data = dune.env, permutations = 999, method = "bray")
#基于bray-curtis距离进行anosim分析
anosim = anosim(dune, dune.env$Management, permutations = 999, distance = "bray")
# 应力值stress,Adonis R2与显著性,Anosim R与显著性
stress_text <- paste("Stress =", round(stress, 4))
adonis_text <- paste(paste("Adonis =", round(adonis$R2, 2)), "**")[1]
anosim_text <- paste(paste("Anosim =", round(anosim$statistic, 2)), "**")
p <- ggplot(df, aes(MDS1, MDS2))+
geom_point(aes(color = Management), size = 5)+
geom_polygon(aes(x = MDS1, y = MDS2, fill = Management, group = Management, color = Management),
alpha = 0.3, linetype = "longdash", linewidth = 1.5)+
theme(plot.margin = unit(rep(1, 4), 'lines'),
panel.border = element_rect(fill = NA, color = "black", size = 0.5, linetype = "solid"),
panel.grid = element_blank(),
panel.background = element_rect(fill = 'white'))+
guides(color = "none", fill = "none")+
ggtitle(paste(paste(stress_text, adonis_text), anosim_text))
ggsave("Figure6C.pdf", p, height = 5.69, width = 7.42)猜你喜欢
iMeta简介 高引文章 高颜值绘图imageGP 网络分析iNAP
iMeta网页工具 代谢组MetOrigin 美吉云乳酸化预测DeepKla
iMeta综述 肠菌菌群 植物菌群 口腔菌群 蛋白质结构预测
10000+:菌群分析 宝宝与猫狗 梅毒狂想曲 提DNA发Nature
一文读懂:宏基因组 寄生虫益处 进化树 必备技能:提问 搜索 Endnote
16S功能预测 PICRUSt FAPROTAX Bugbase Tax4Fun
生物科普: 肠道细菌 人体上的生命 生命大跃进 细胞暗战 人体奥秘
写在后面
为鼓励读者交流快速解决科研困难,我们建立了“宏基因组”讨论群,己有国内外6000+ 科研人员加入。请添加主编微信meta-genomics带你入群,务必备注“姓名-单位-研究方向-职称/年级”。高级职称请注明身份,另有海内外微生物PI群供大佬合作交流。技术问题寻求帮助,首先阅读《如何优雅的提问》学习解决问题思路,仍未解决群内讨论,问题不私聊,帮助同行。
点击阅读原文,跳转最新文章目录阅读























 1559
1559

 被折叠的 条评论
为什么被折叠?
被折叠的 条评论
为什么被折叠?








